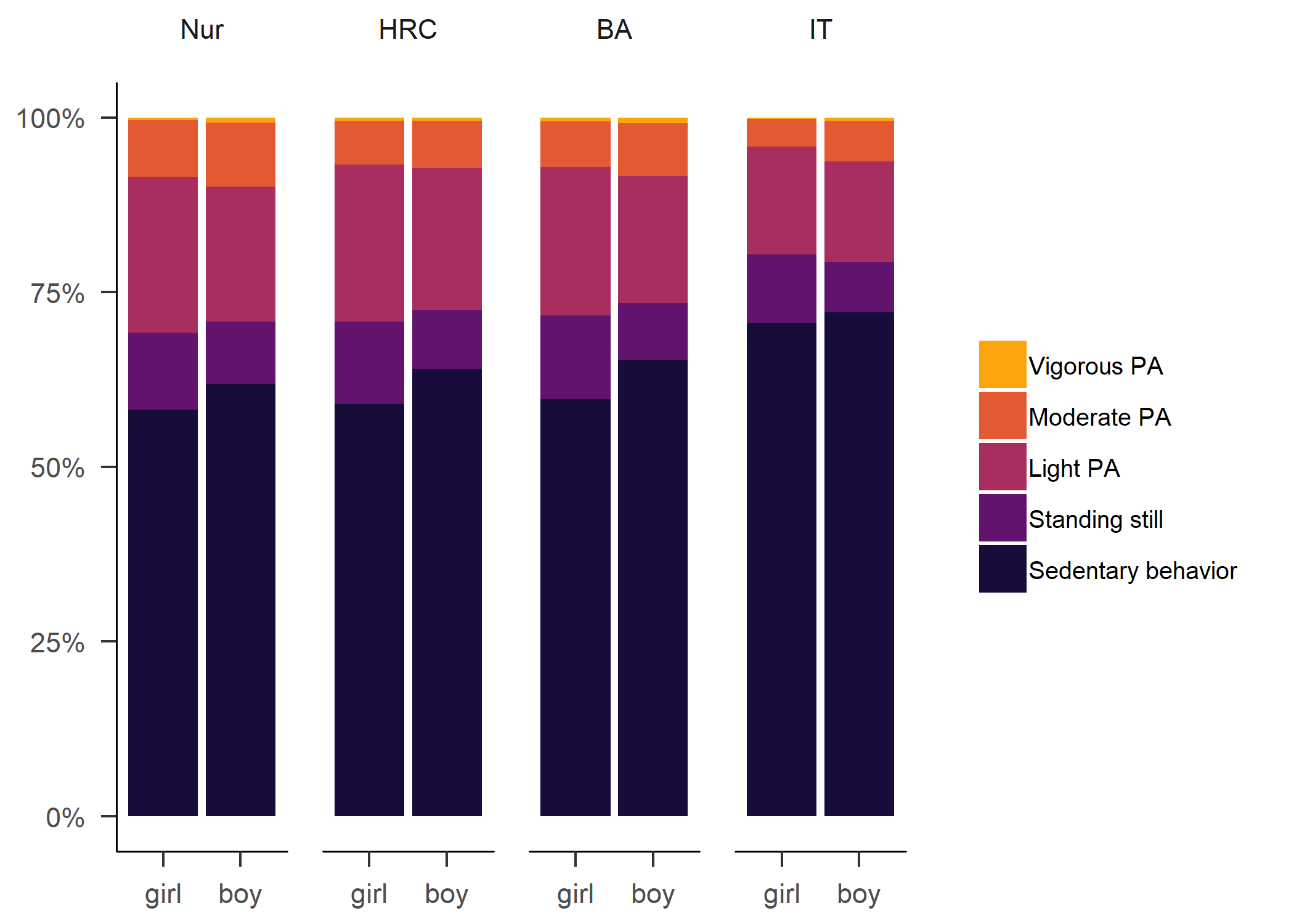
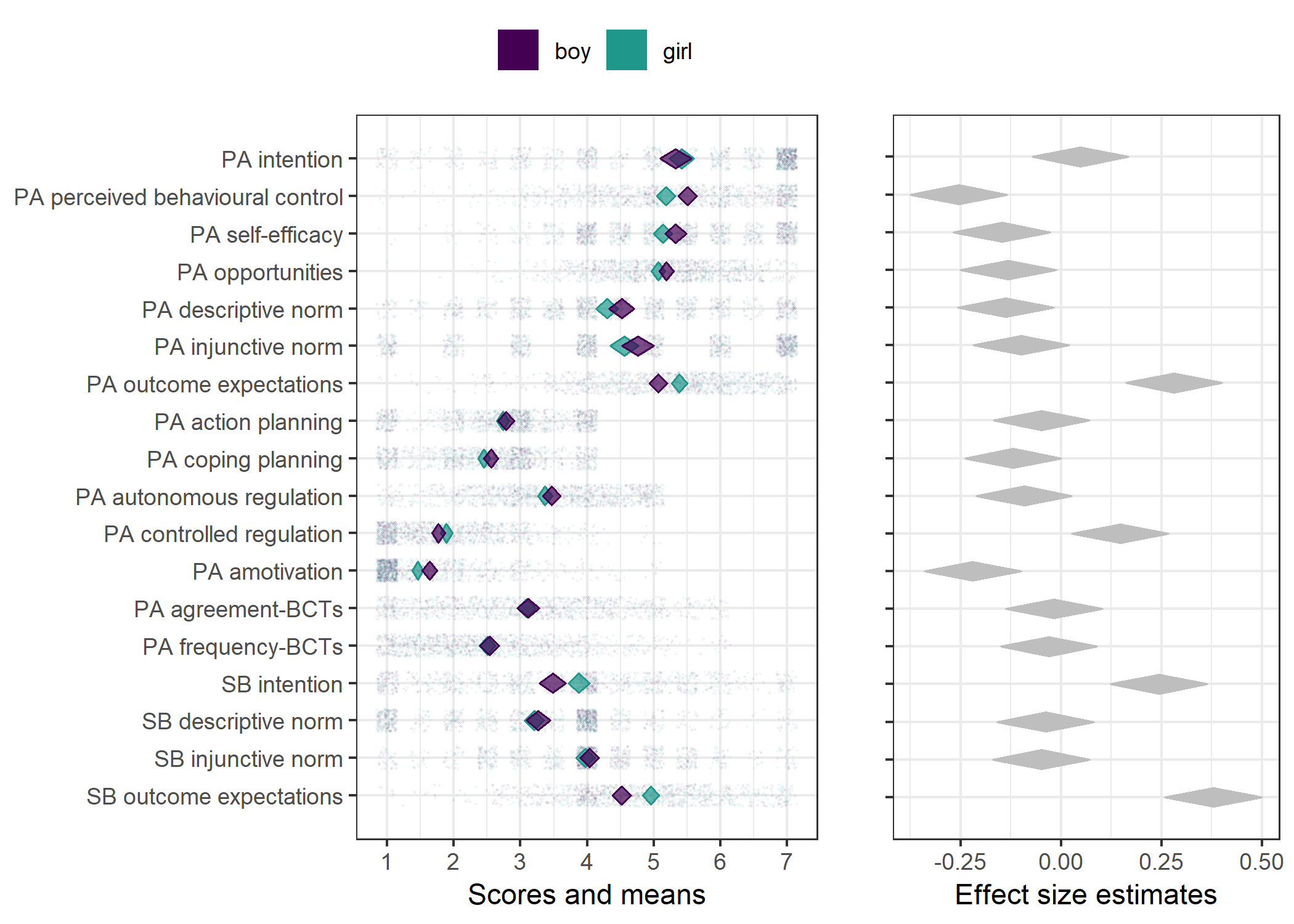
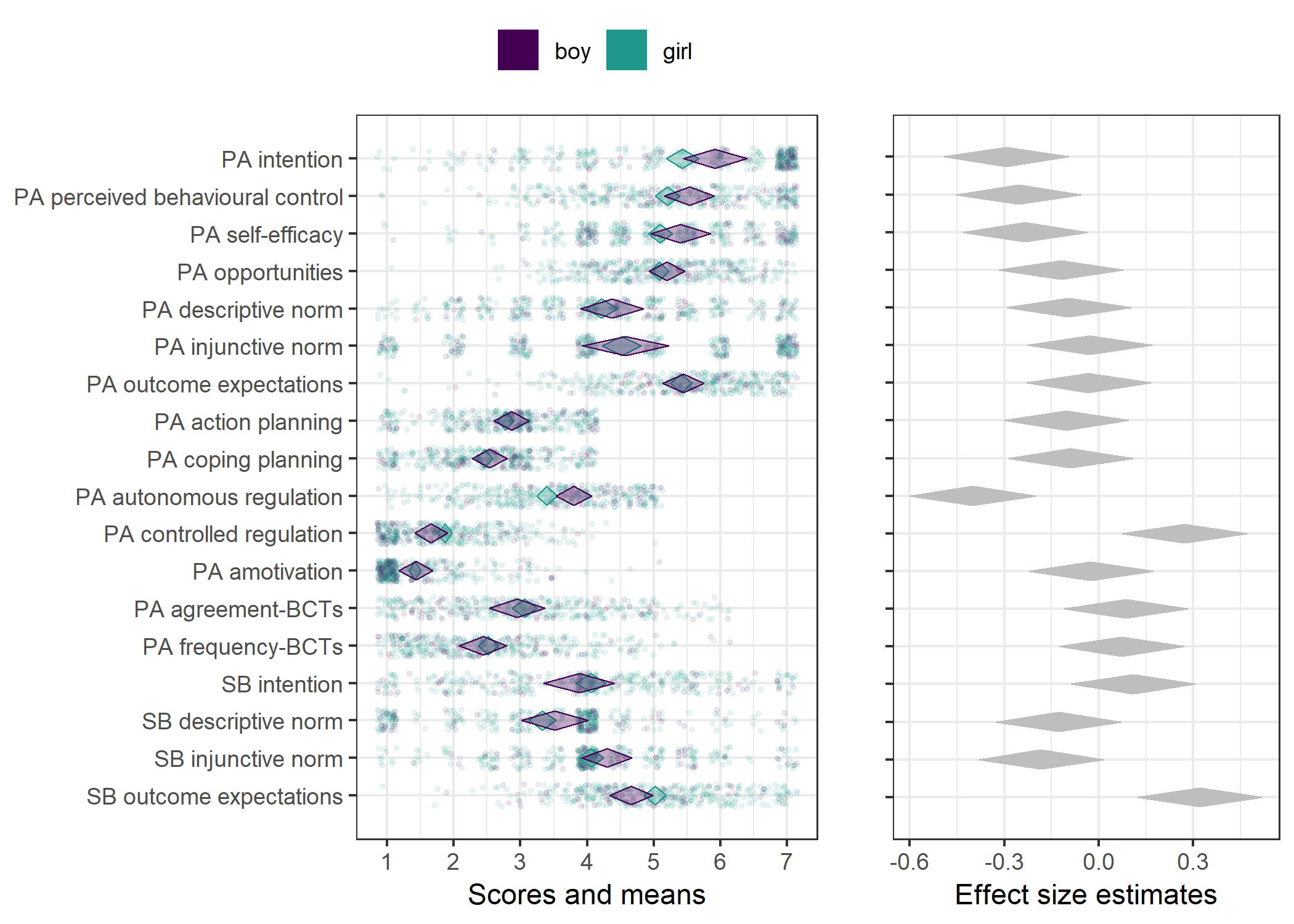
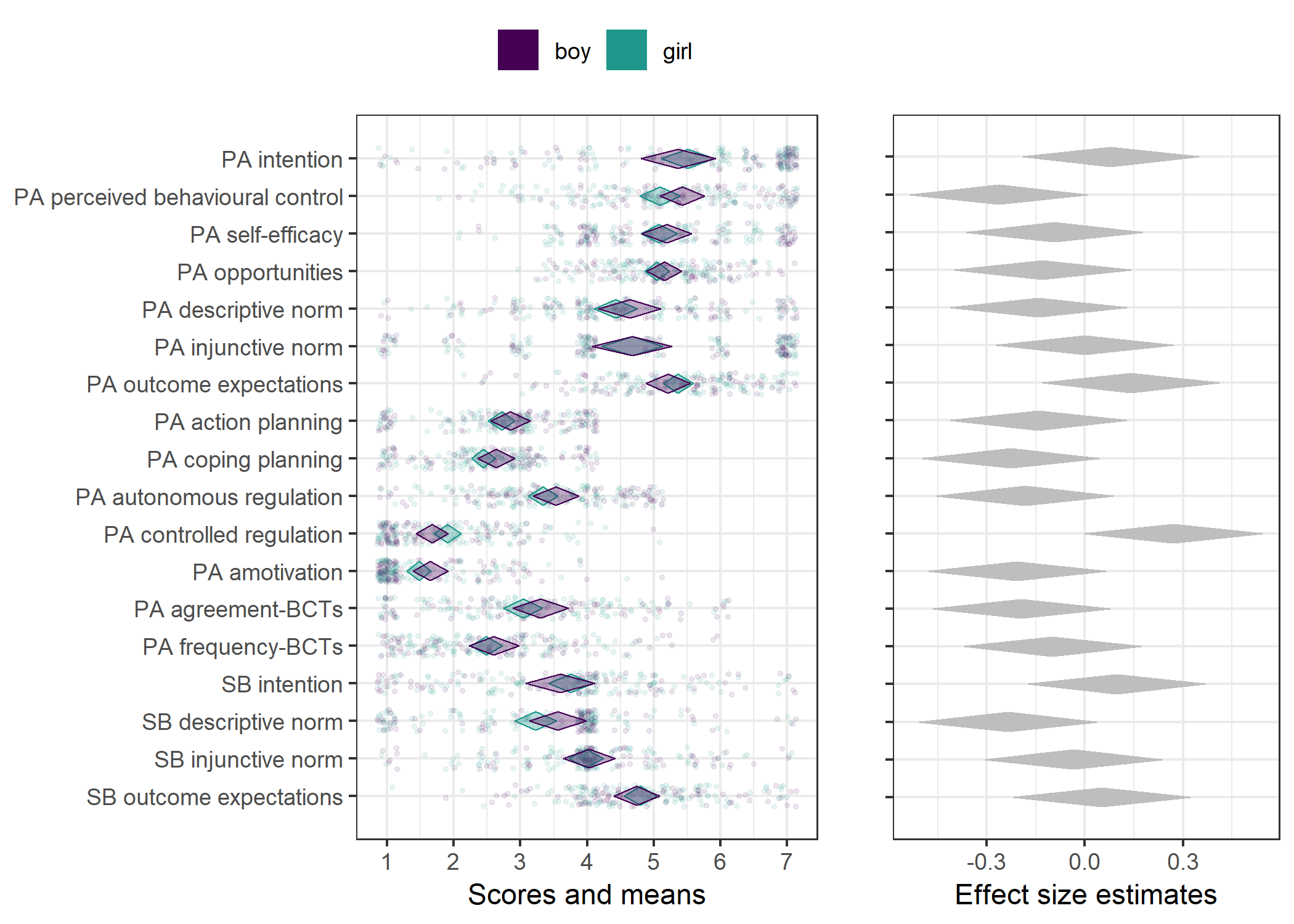
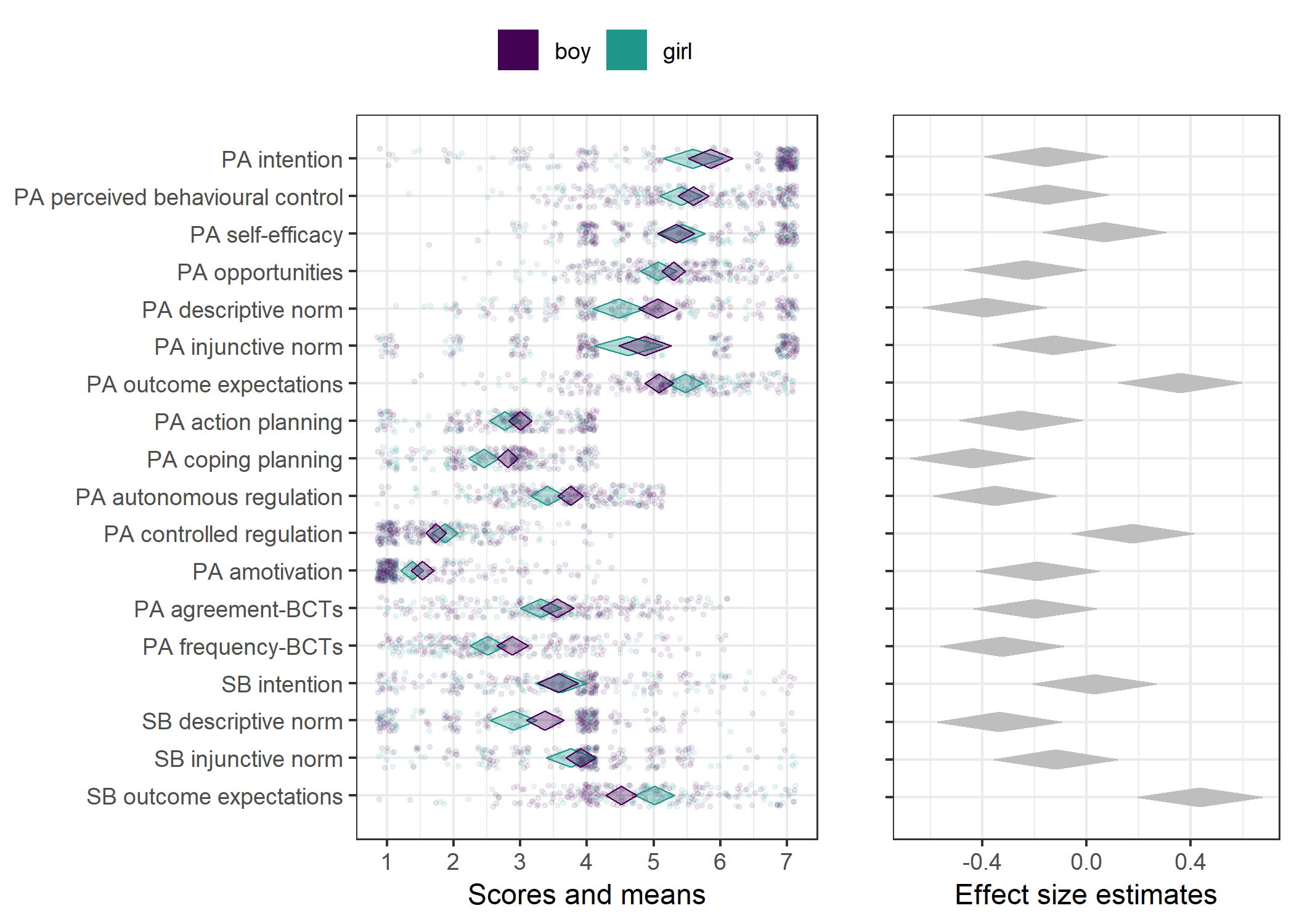
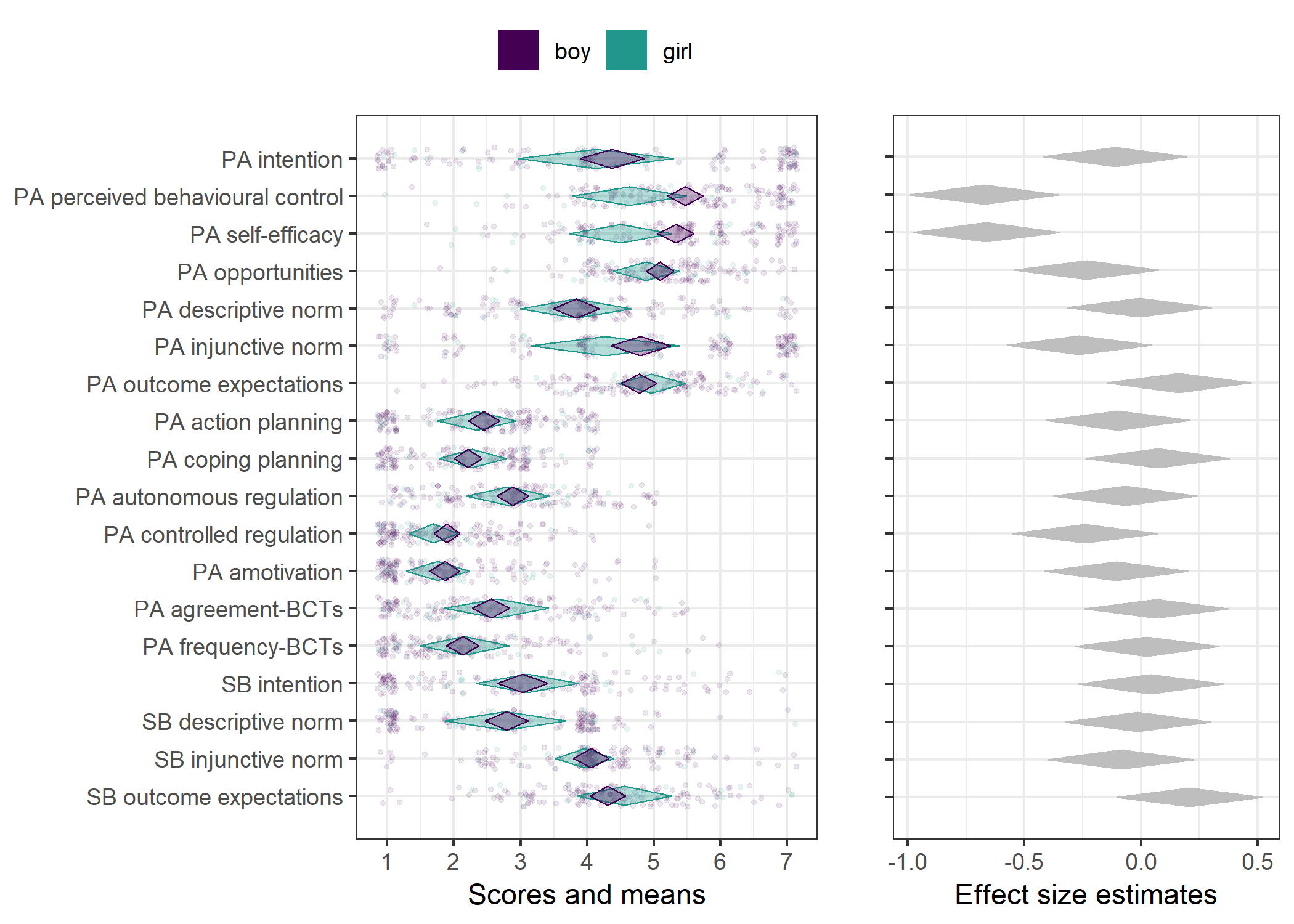
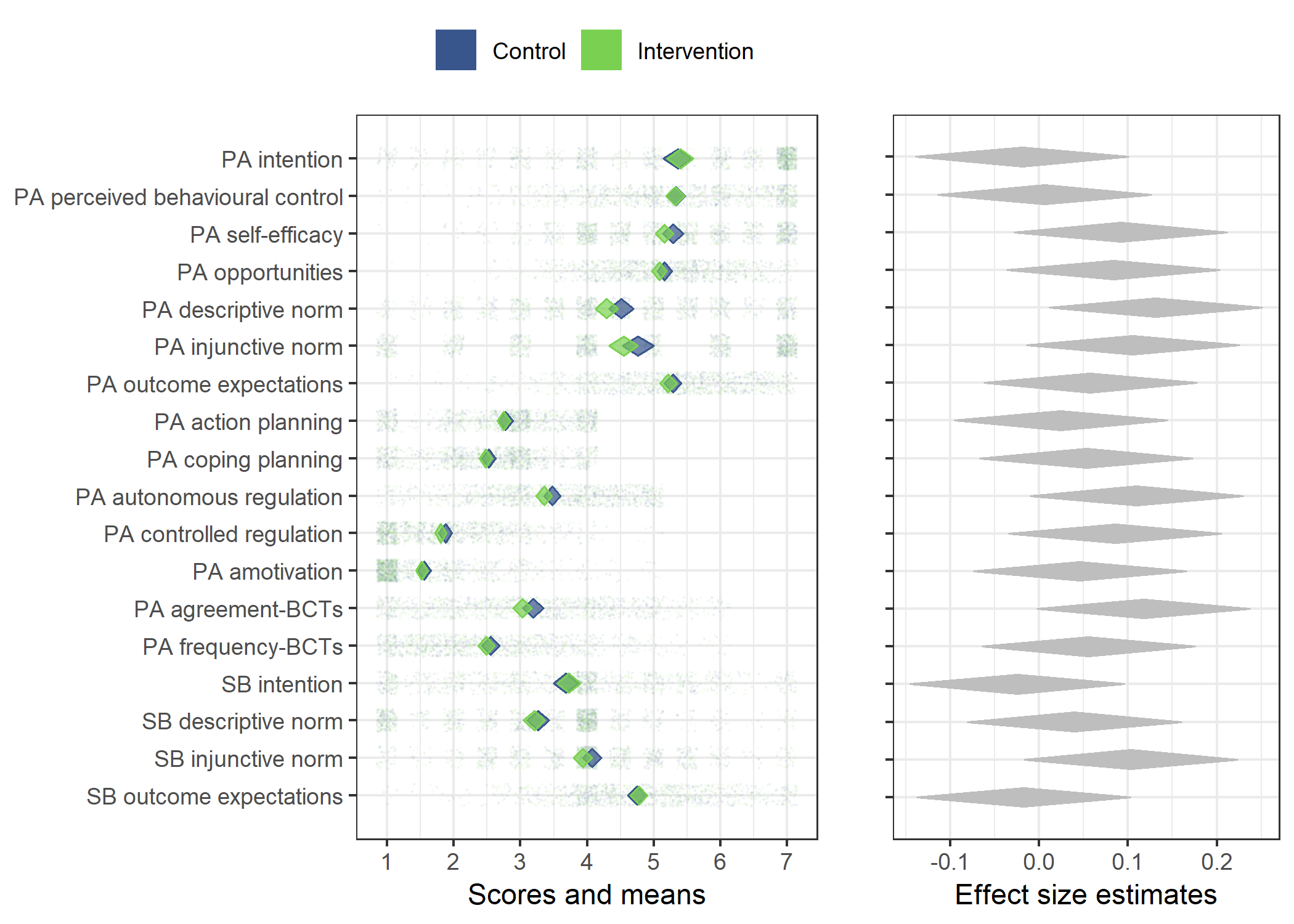
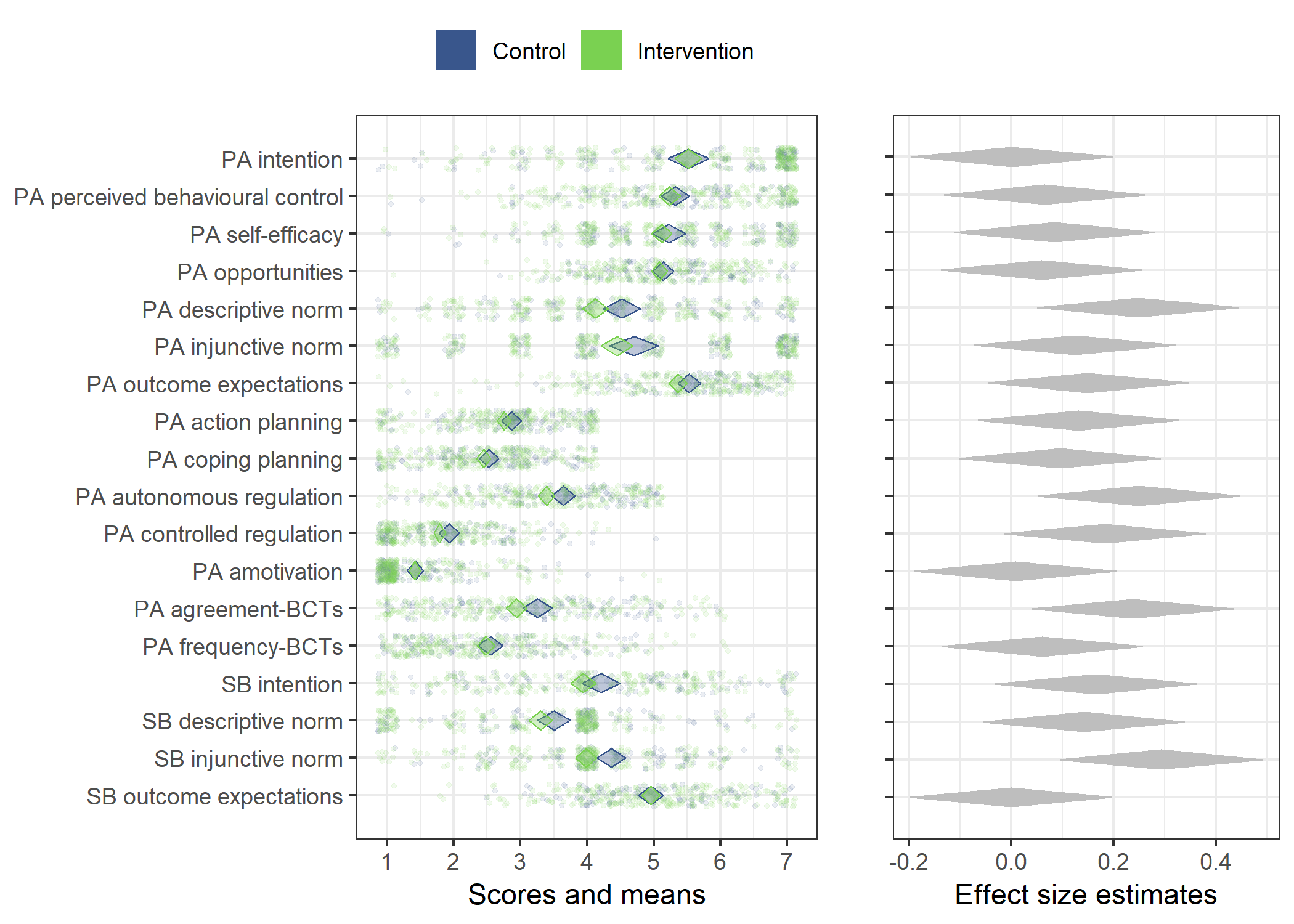
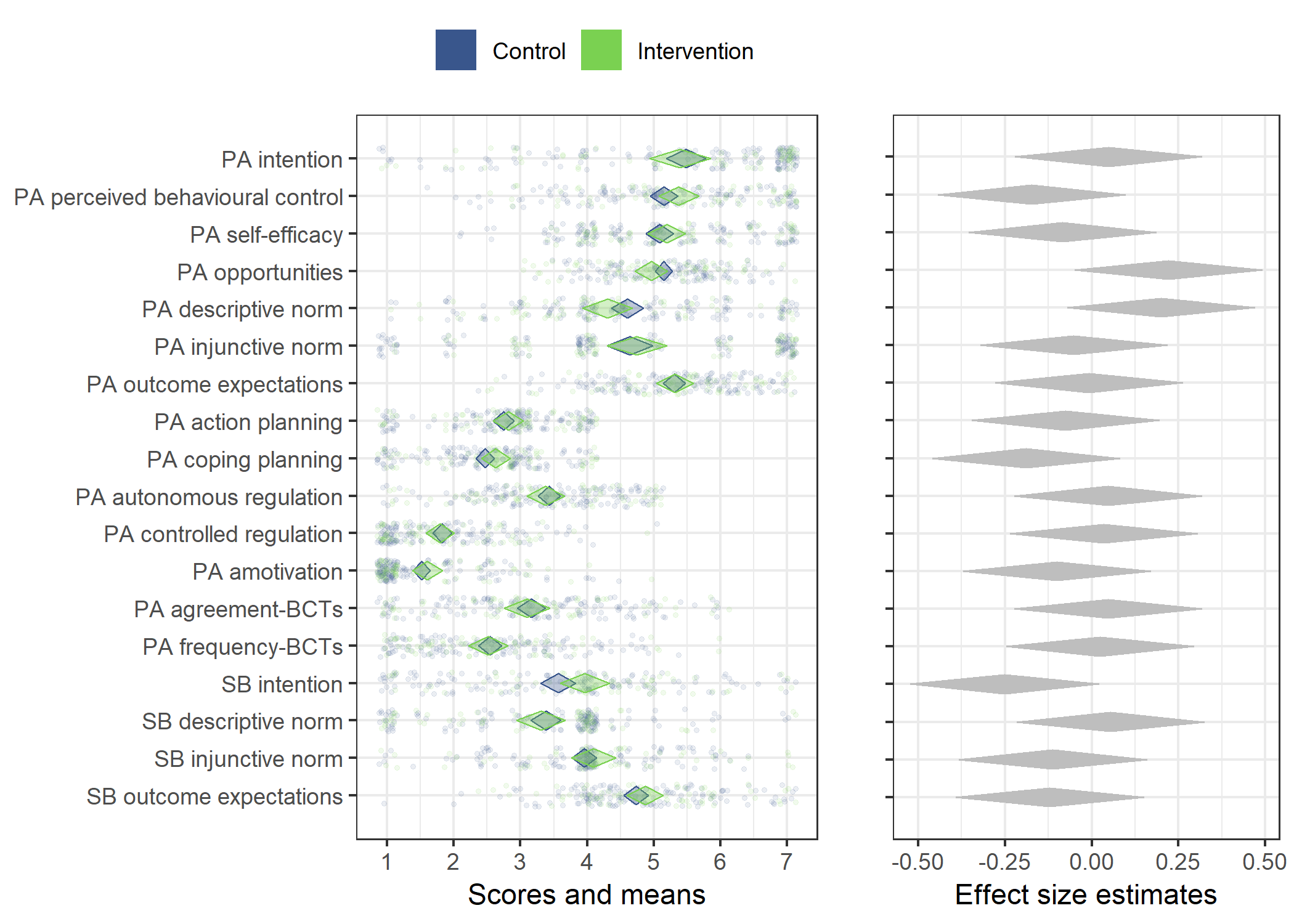
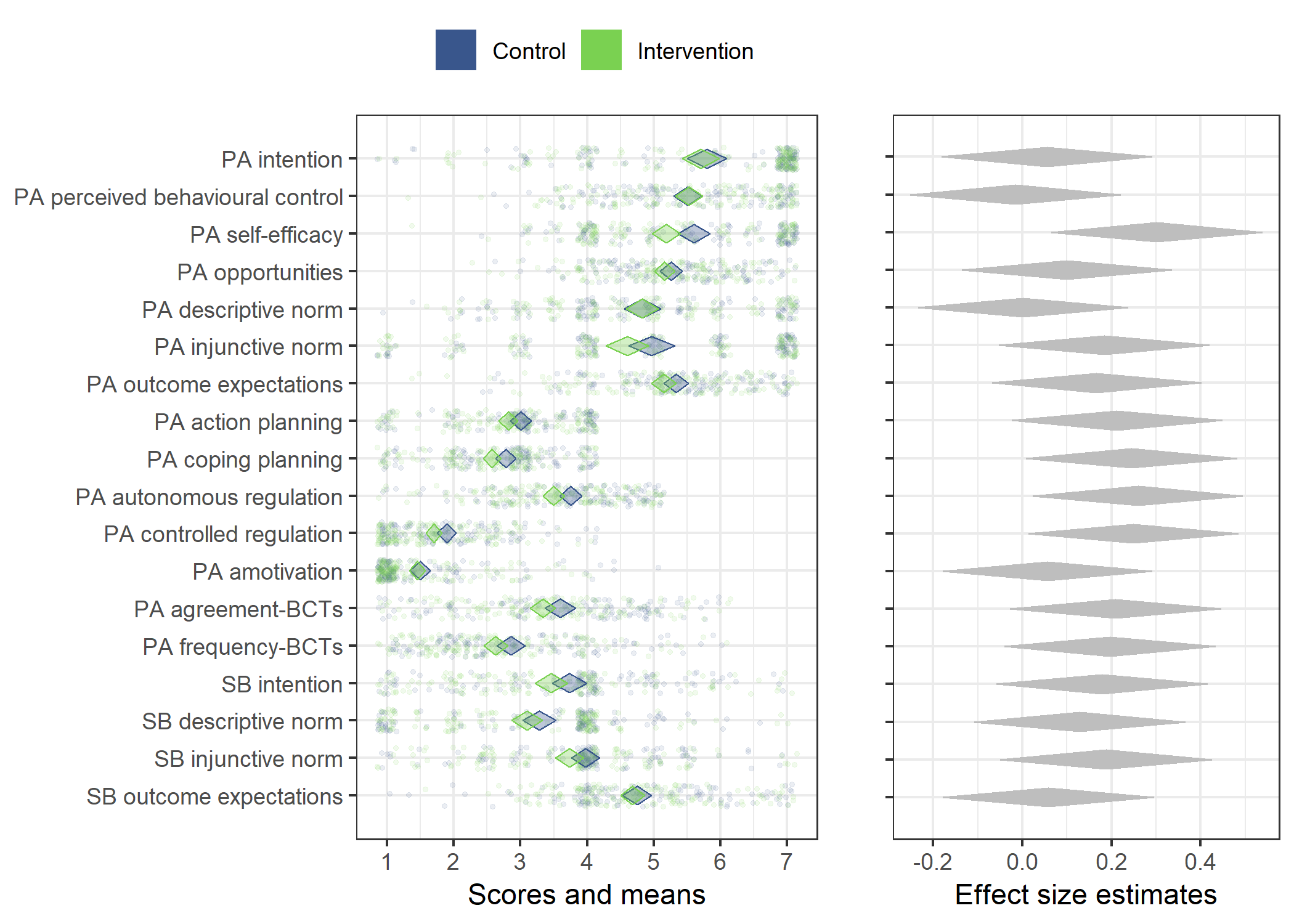
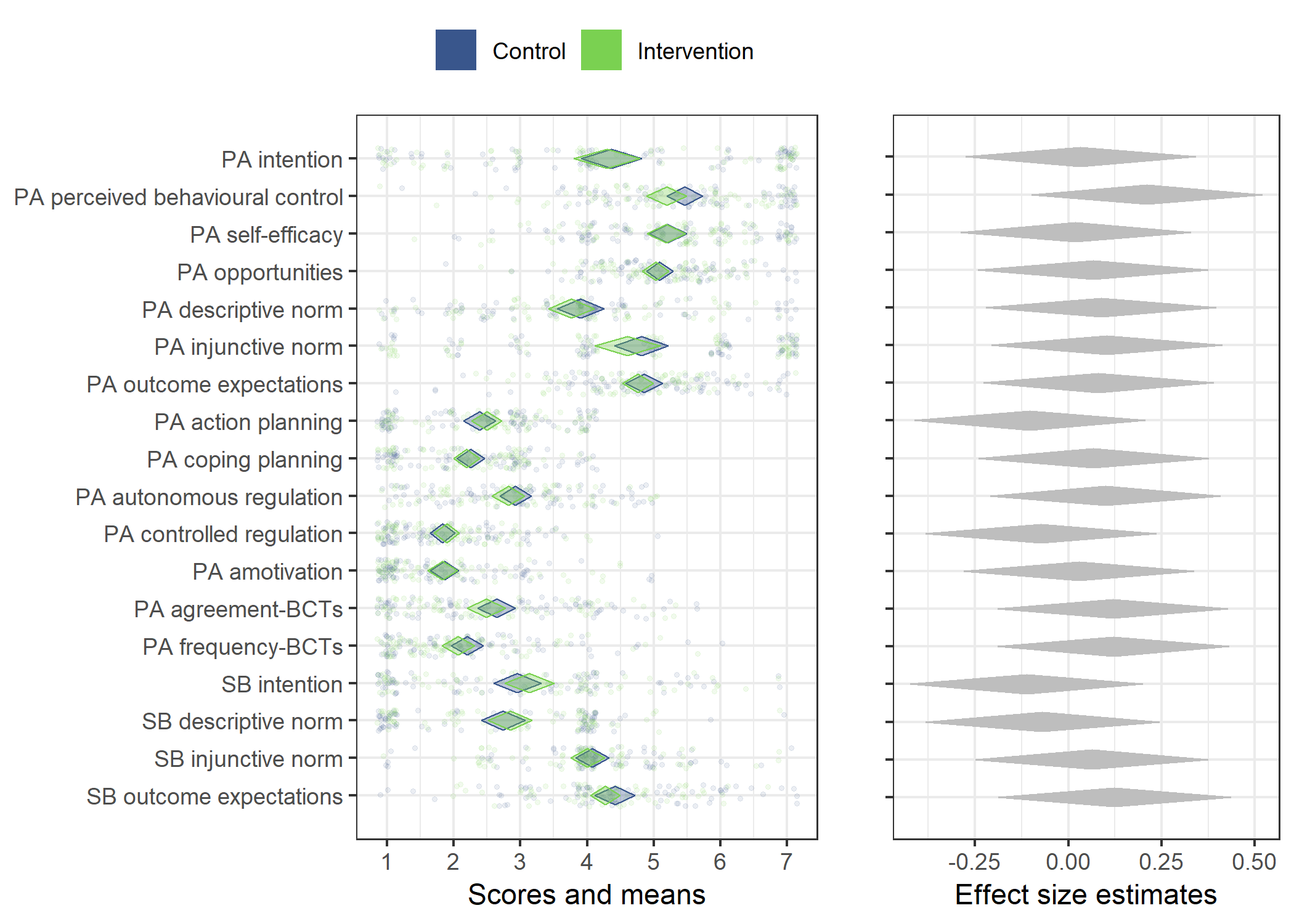
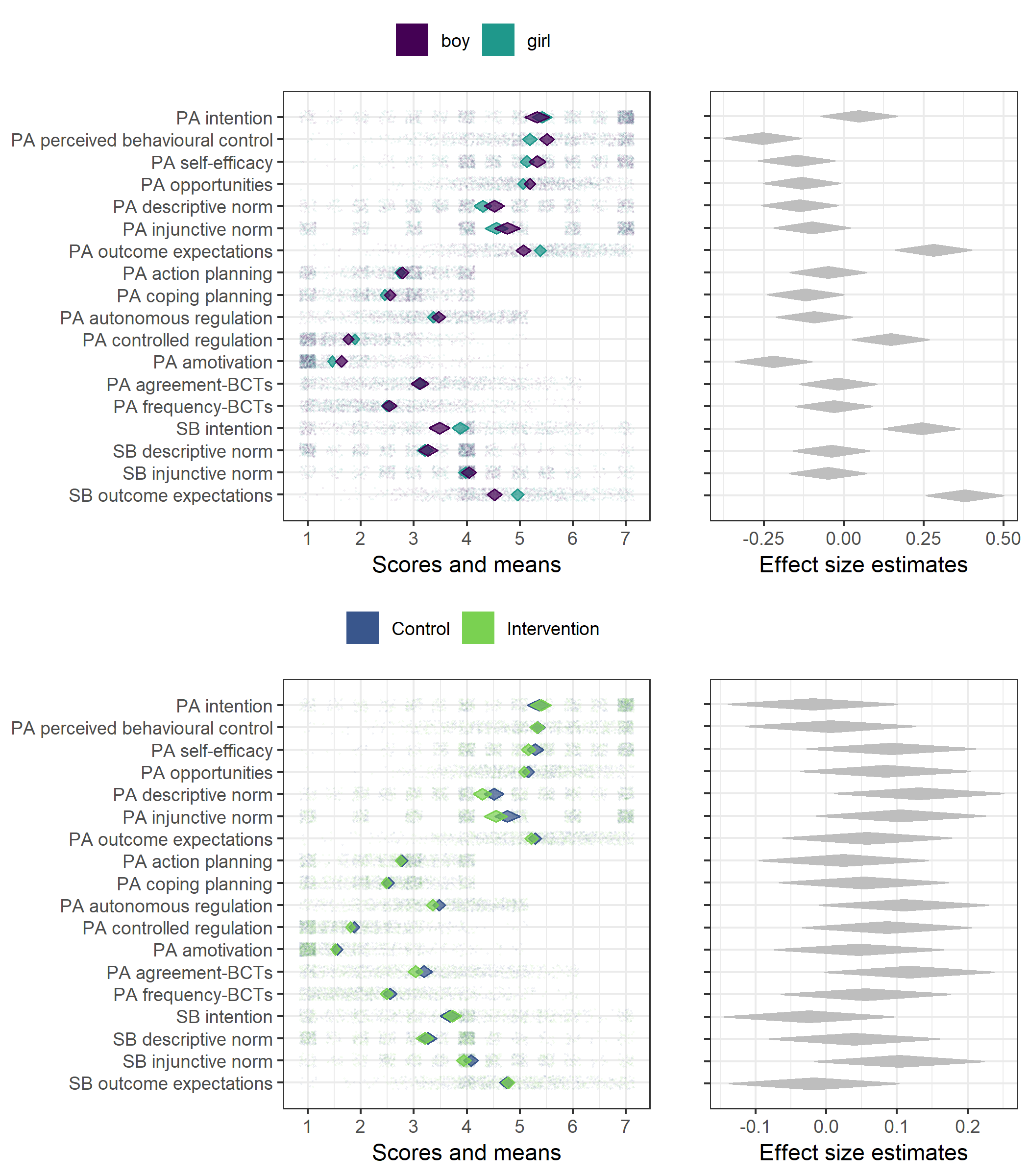
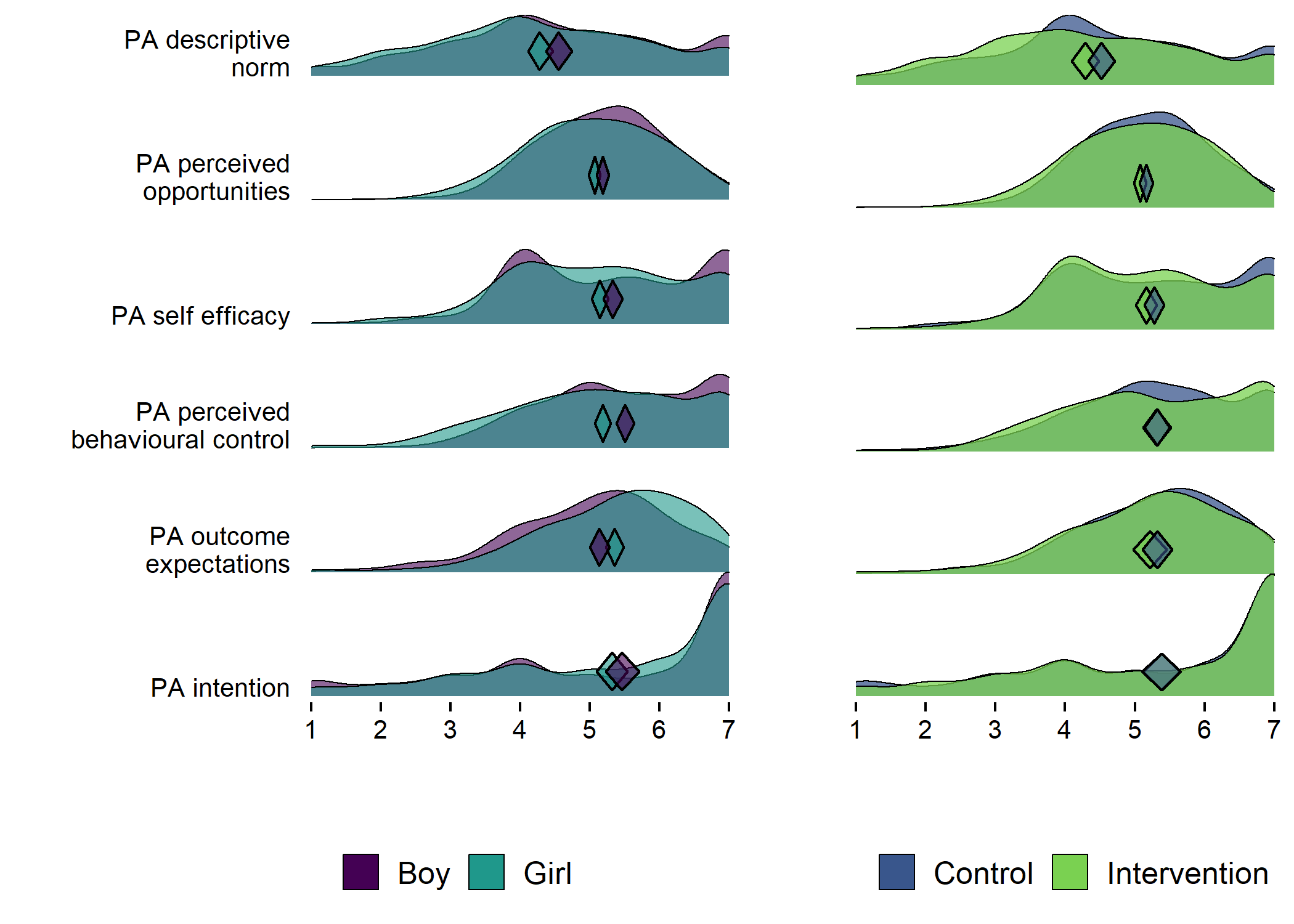
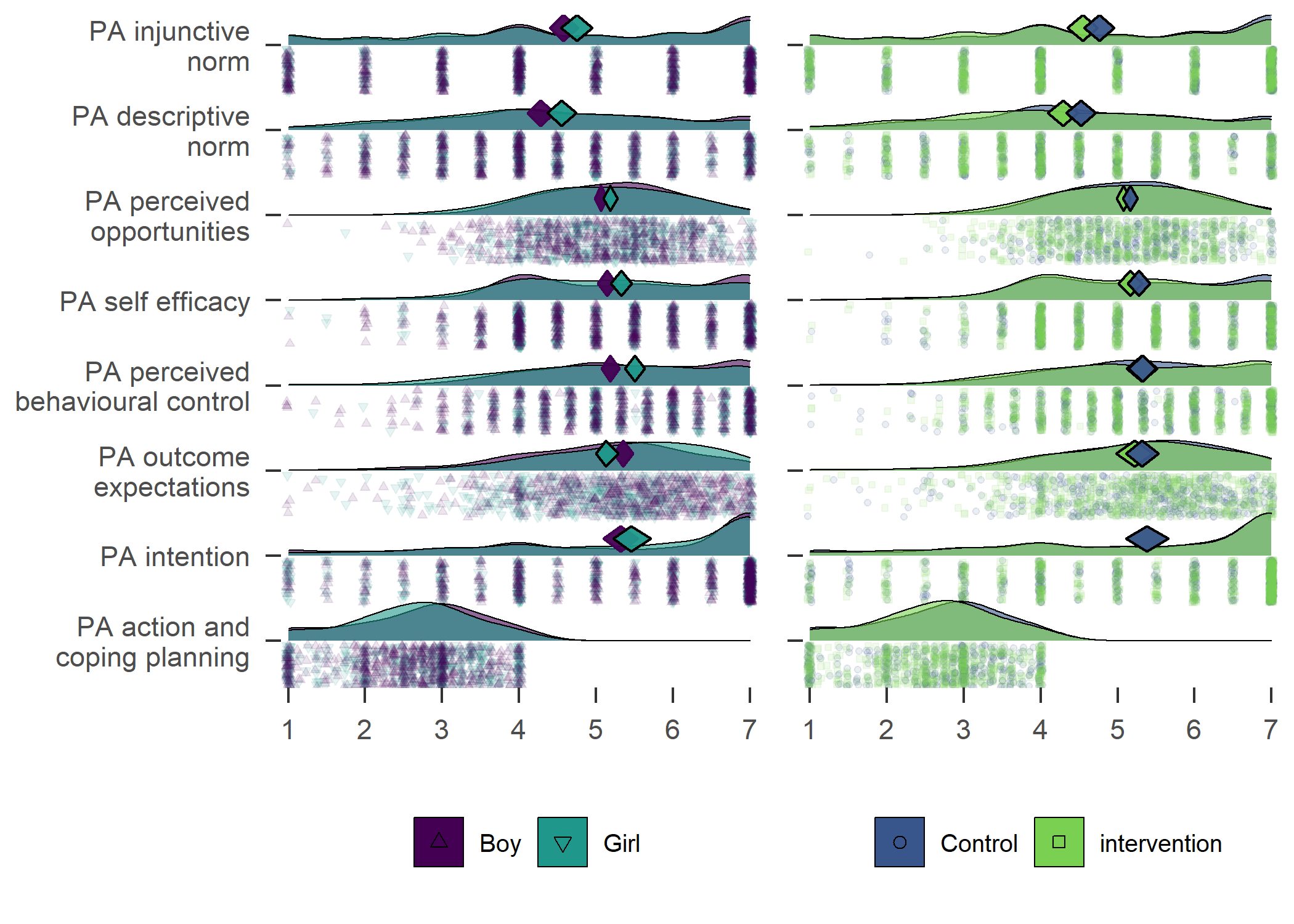
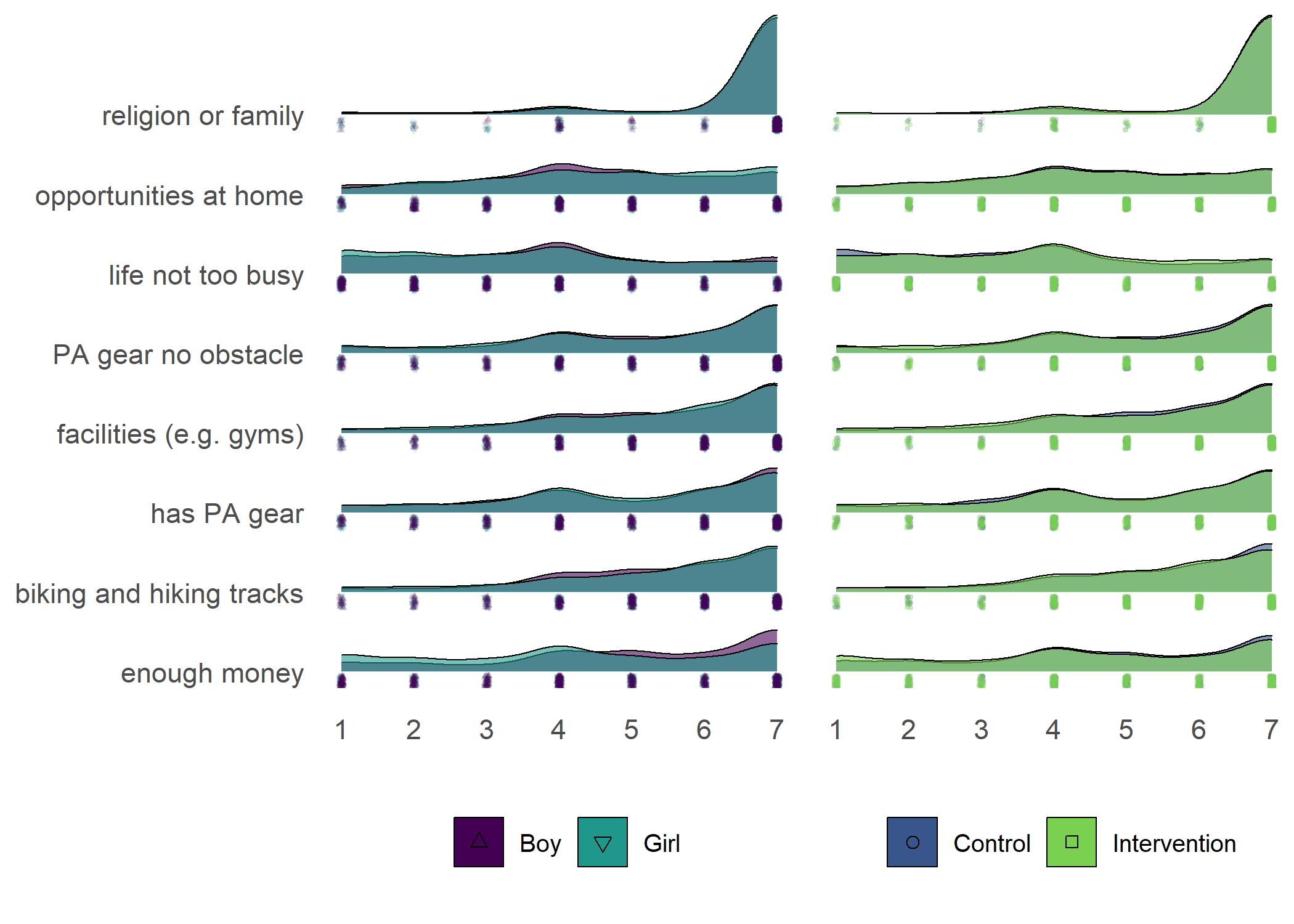
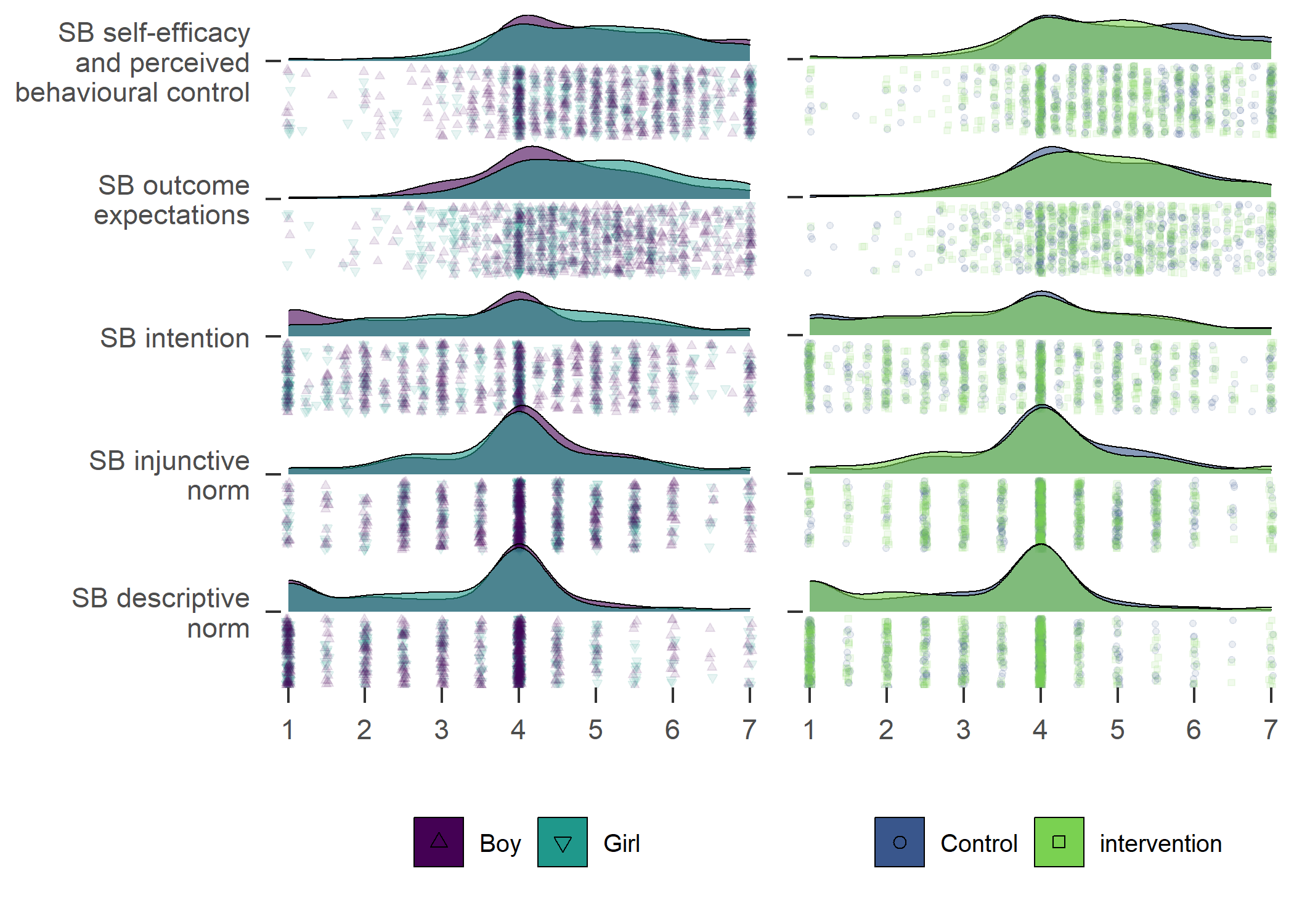
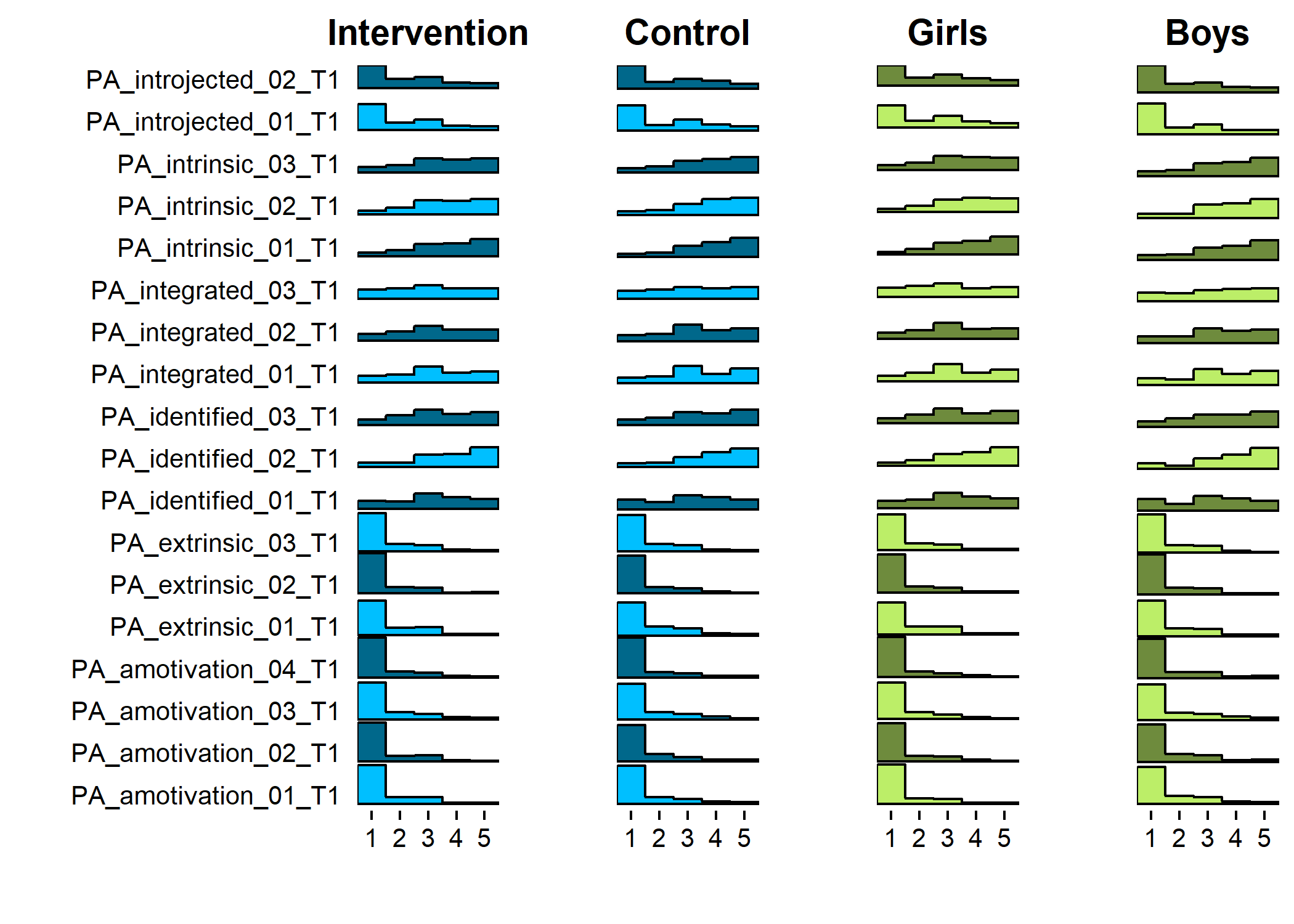
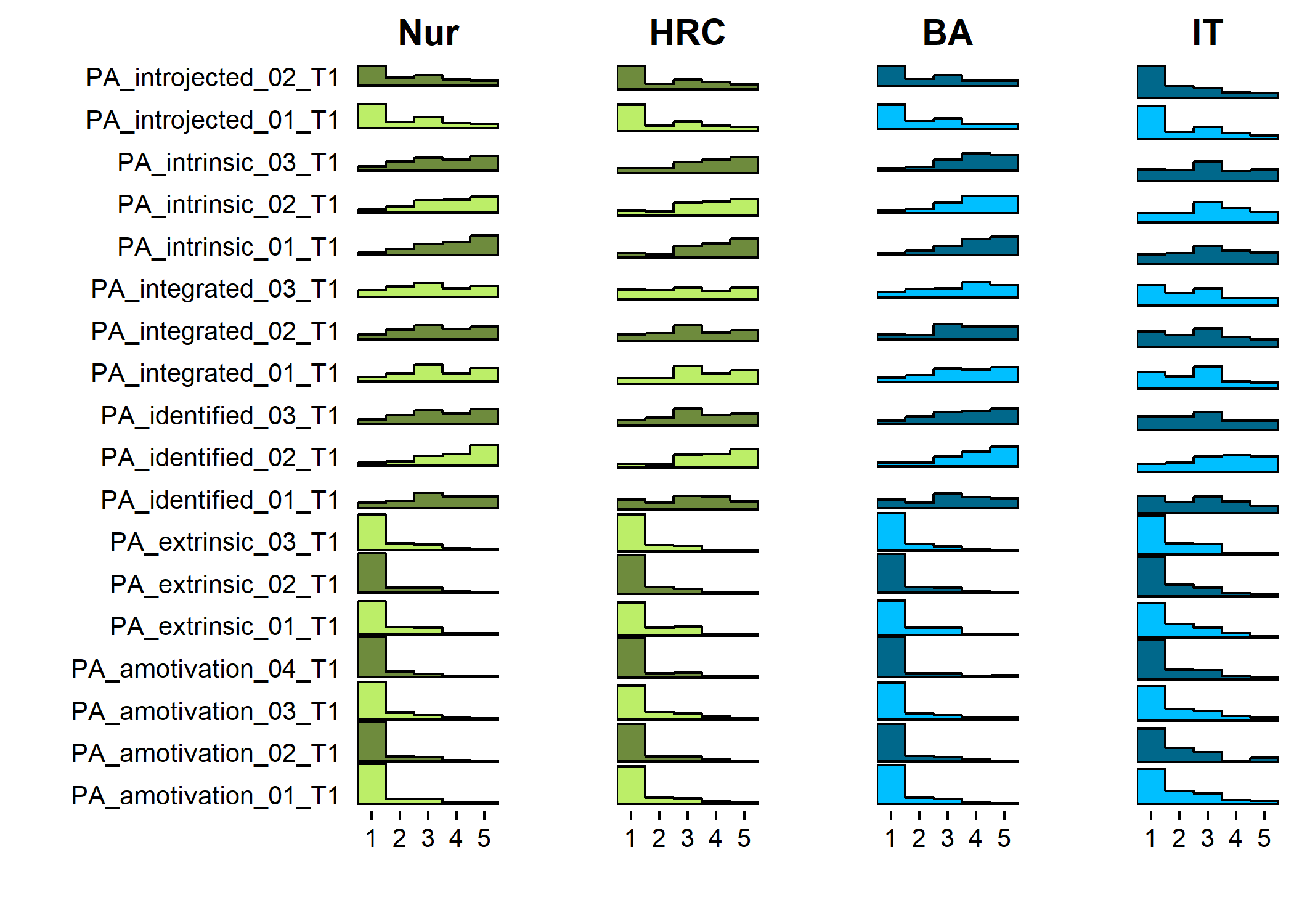
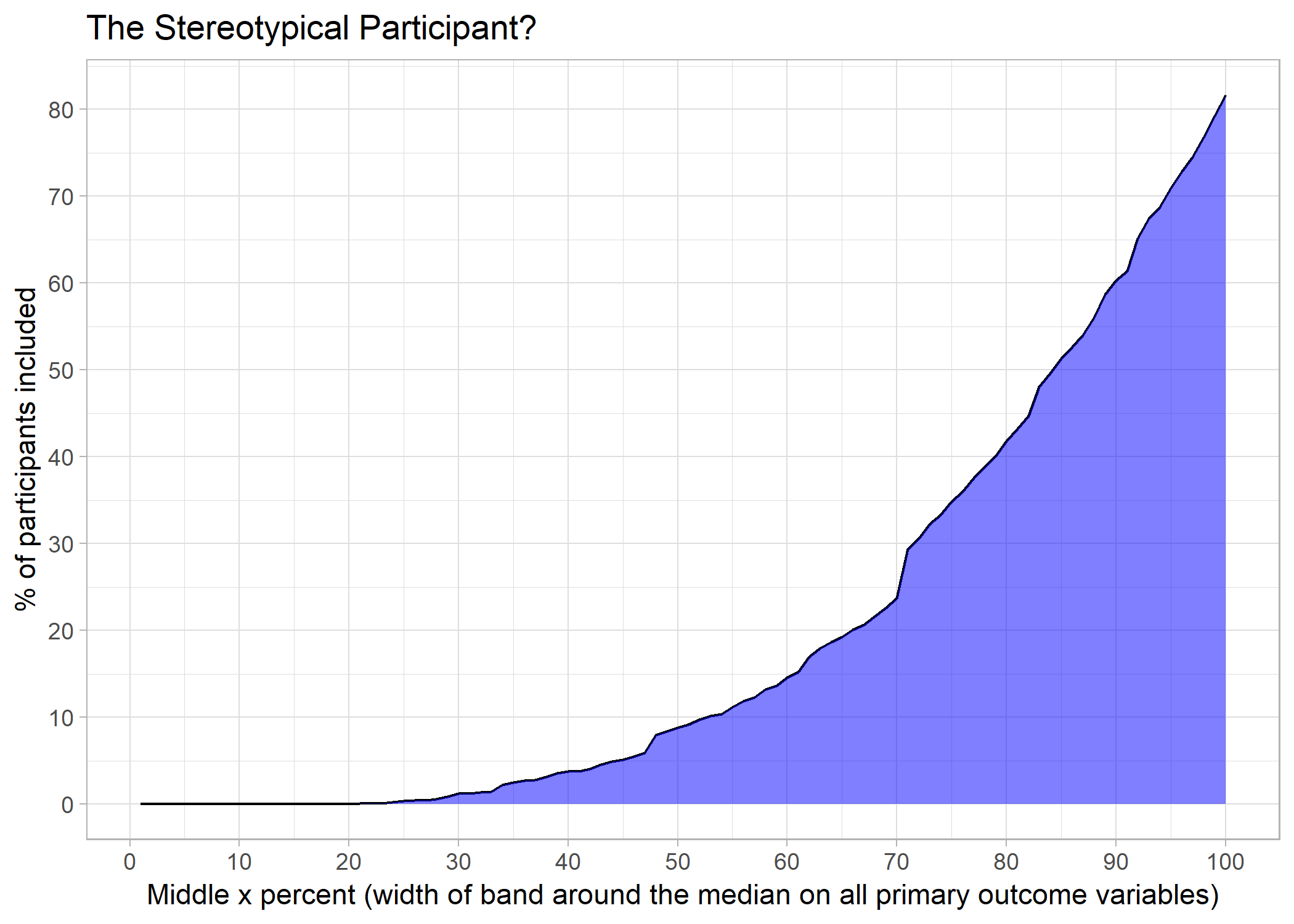
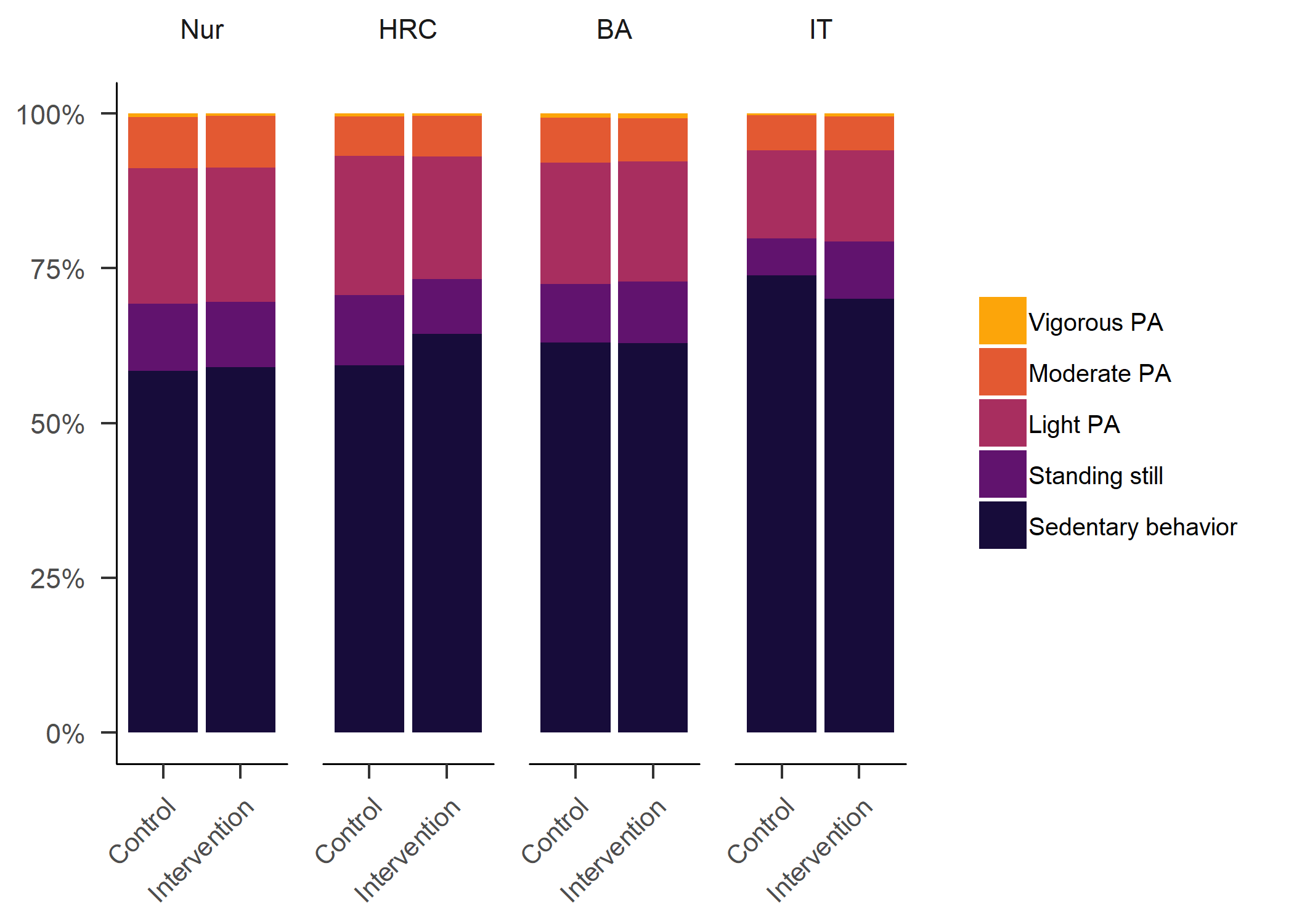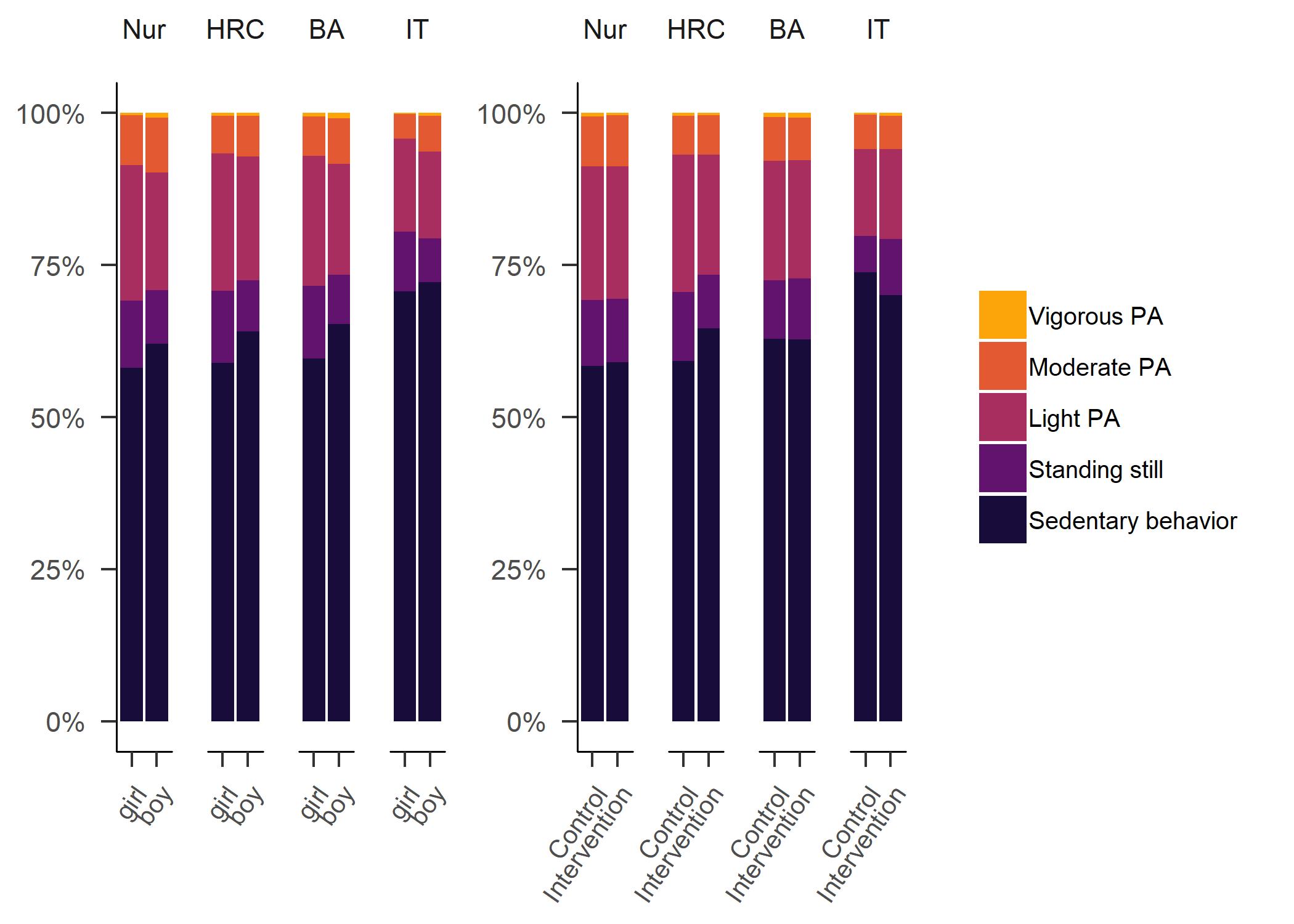Primary outcome variables
[code chunk below transforms MVPA, SB and wear time to hours for ease of interpretation]
d <- d %>% dplyr::mutate(
mvpaAccelerometer_T1 = mvpaAccelerometer_T1 / 60,
# mvpaAccelerometer_T3 = mvpaAccelerometer_T3 / 60,
weartimeAccelerometer_T1 = weartimeAccelerometer_T1 / 60,
# weartimeAccelerometer_T3 = weartimeAccelerometer_T3 / 60,
sitLieAccelerometer_T1 = sitLieAccelerometer_T1 / 60
# sitLieAccelerometer_T3 = sitLieAccelerometer_T3 / 60
)
df <- df %>% dplyr::mutate(
mvpaAccelerometer_T1 = mvpaAccelerometer_T1 / 60,
# mvpaAccelerometer_T3 = mvpaAccelerometer_T3 / 60,
weartimeAccelerometer_T1 = weartimeAccelerometer_T1 / 60,
# weartimeAccelerometer_T3 = weartimeAccelerometer_T3 / 60,
sitLieAccelerometer_T1 = sitLieAccelerometer_T1 / 60
# sitLieAccelerometer_T3 = sitLieAccelerometer_T3 / 60
)Self-reported vs. accelerometer-measured MVPA
ggstatsplot::ggscatterstats(
data = na.omit(dplyr::select(df, mvpaAccelerometer_T1, padaysLastweek_T1)),
x = mvpaAccelerometer_T1,
y = padaysLastweek_T1,
type = "robust", # type of test that needs to be run
results.subtitle = FALSE,
xlab = "Accelerometer-measured MVPA hours (following week)", # label for x axis
ylab = "Self-reported days of >30min MVPA (previous week)", # label for y axis
line.color = "black", # changing regression line colour line
title = "", # title text for the plot
# caption = expression( # caption text for the plot
# paste(italic("Note"), ": this is a demo")
# ),
marginal.type = "histogram", # type of marginal distribution to be displayed
xfill = "blue", # colour fill for x-axis marginal distribution
yfill = "red", # colour fill for y-axis marginal distribution
centrality.para = "median" # which type of intercept line is to be displayed
# width.jitter = 0, # amount of horizontal jitter for data points
# height.jitter = 0 # amount of vertical jitter for data points
)
## Warning: This plot can't be further modified with `ggplot2` functions.
## In case you want a `ggplot` object, set `marginal = FALSE`.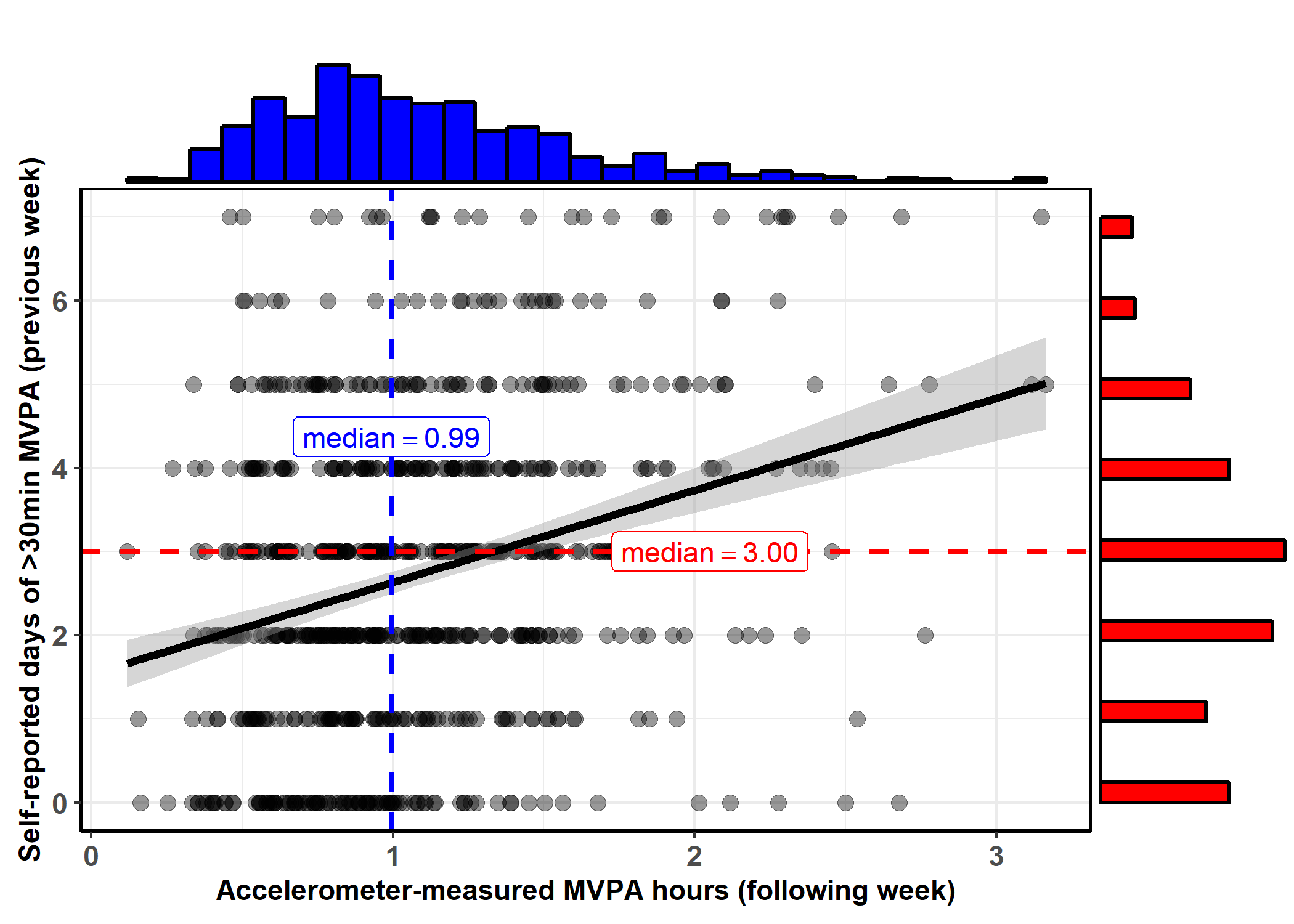
df %>% ggplot2::ggplot(aes(x = mvpaAccelerometer_T1, y = padaysLastweek_T1)) + geom_point() + geom_smooth(method = "loess") +
# geom_vline(xintercept = 120, col = "red") +
labs(x = "accelerometer-measured MVPA hours, following week", y = "self-reported days of 30+ min MVPA, last week")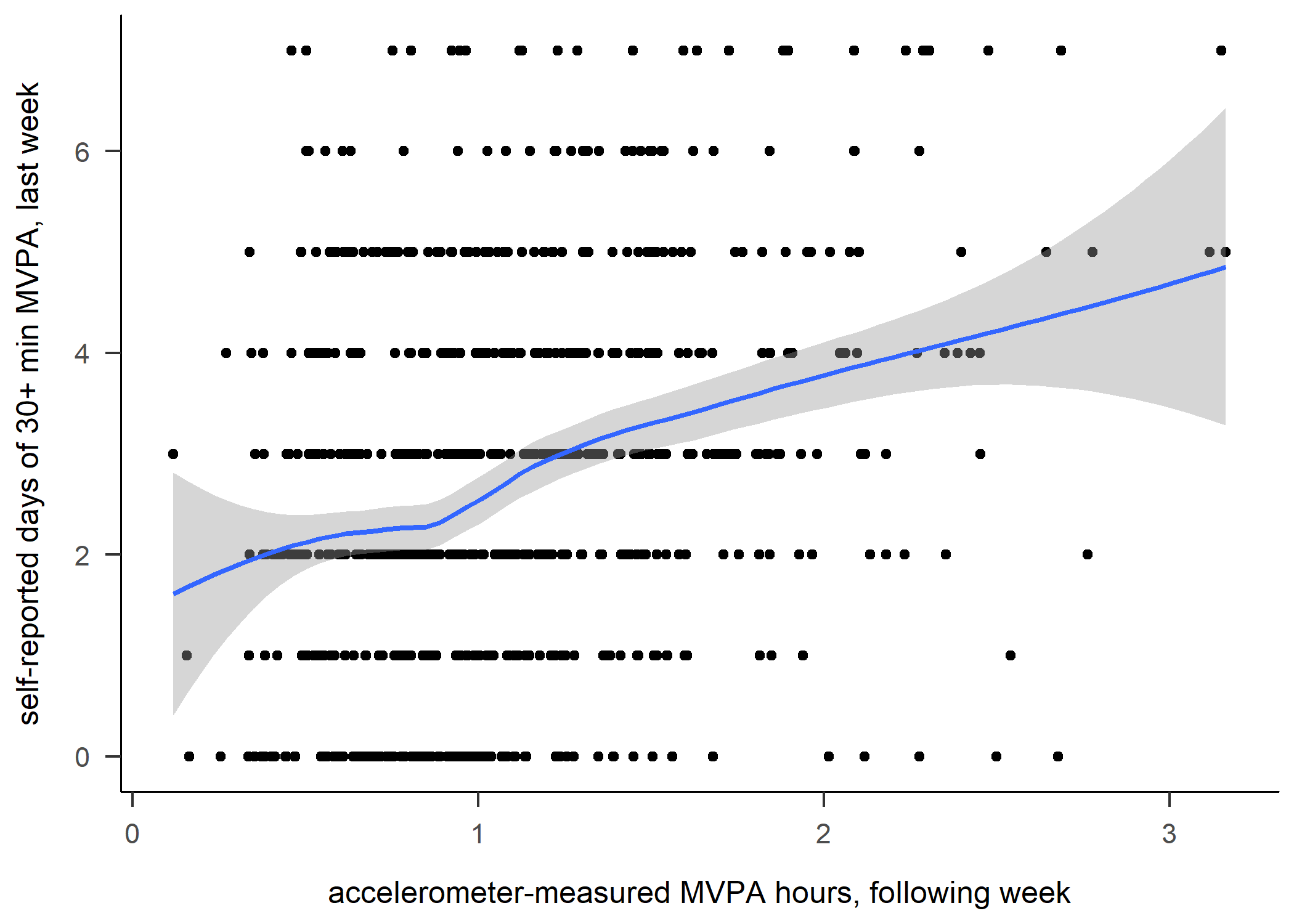
corrgram::corrgram(data.frame("Self-rep PA" = df$padaysLastweek_T1,
"Accelerometer PA" = df$mvpaAccelerometer_T1,
"Sedentary time" = df$sitLieAccelerometer_T1,
"Interrupted sitting" = df$sitBreaksAccelerometer_T1),
lower.panel = corrgram::panel.pie, upper.panel = corrgram::panel.conf)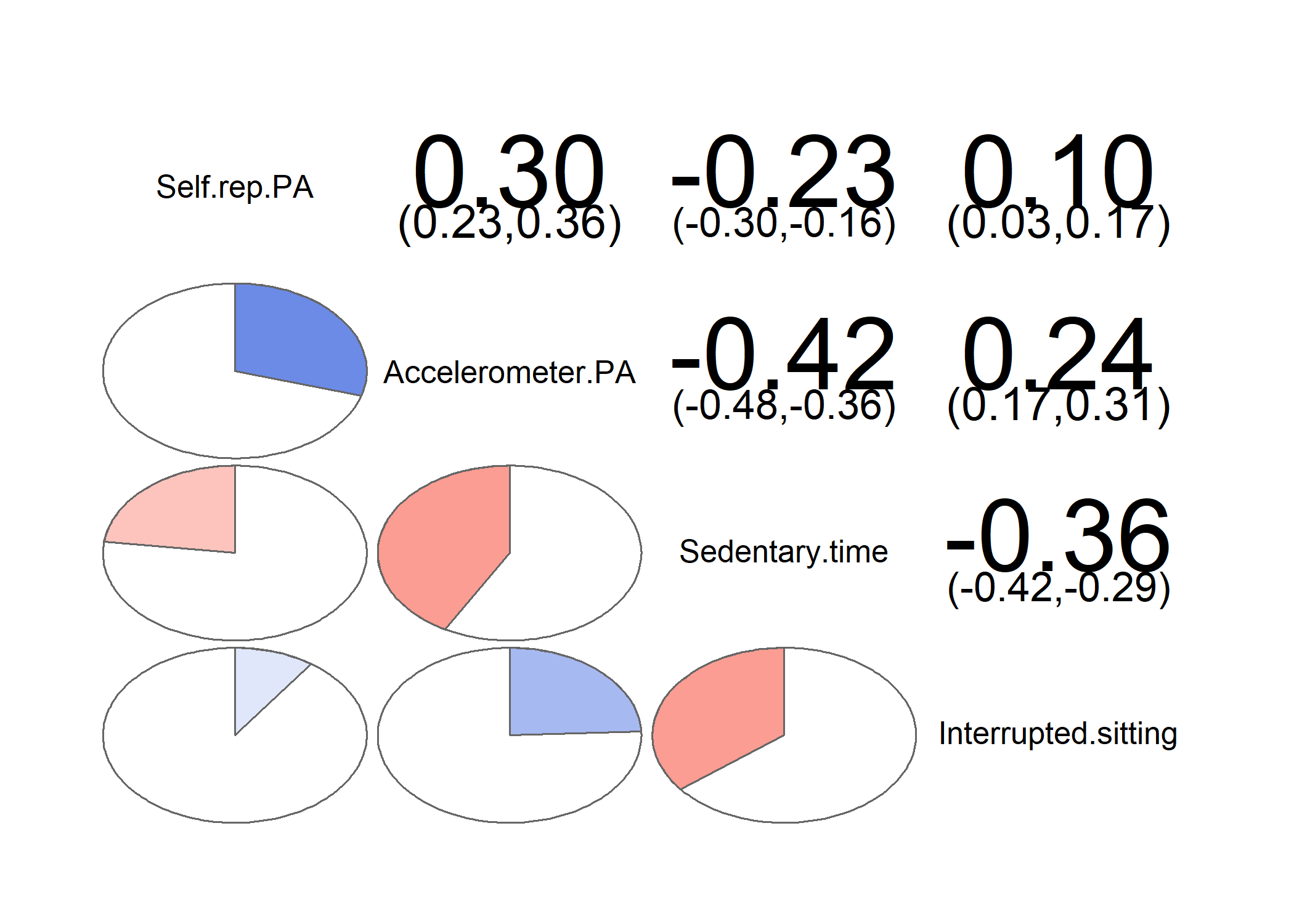
Accelerometer weartime
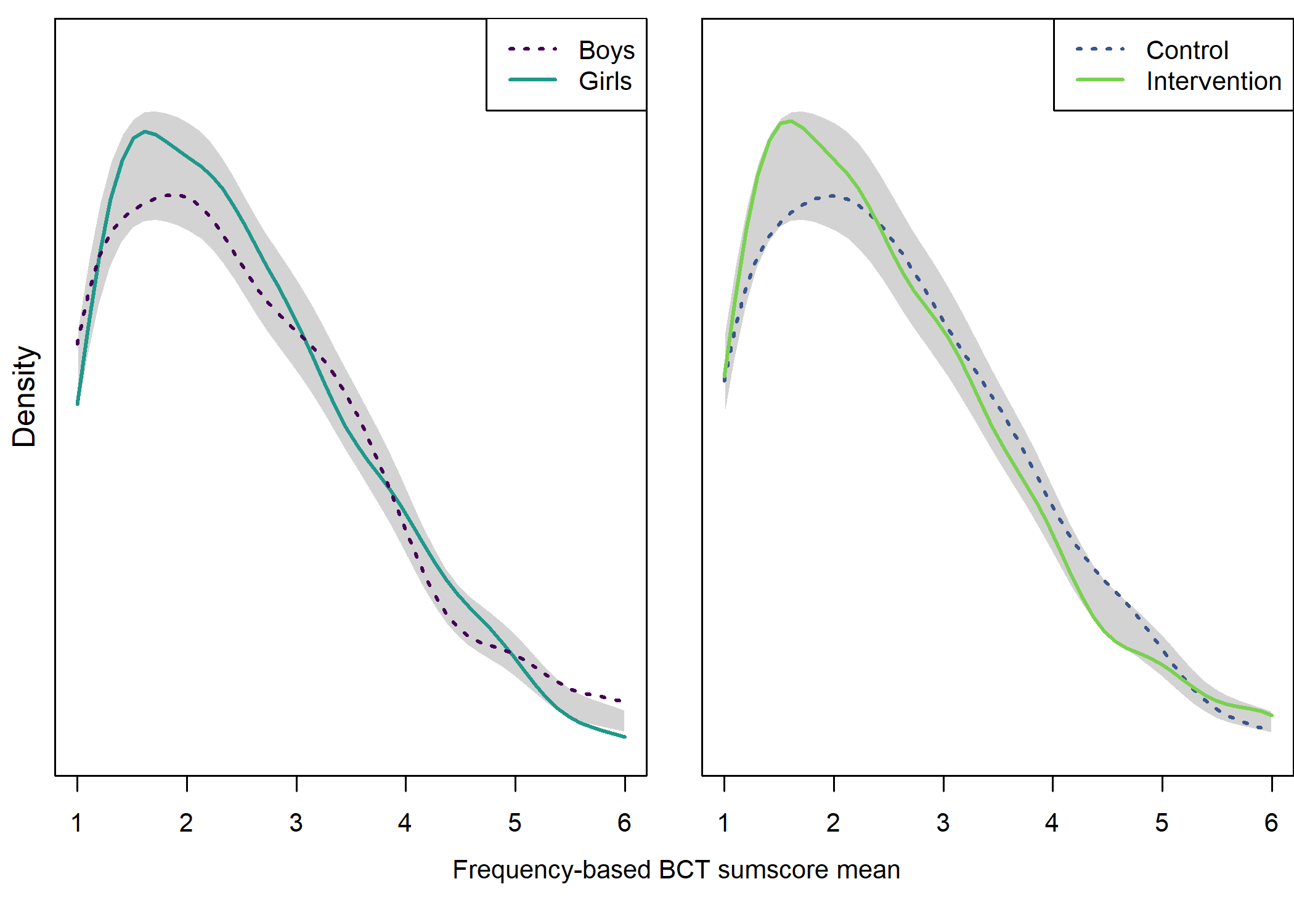
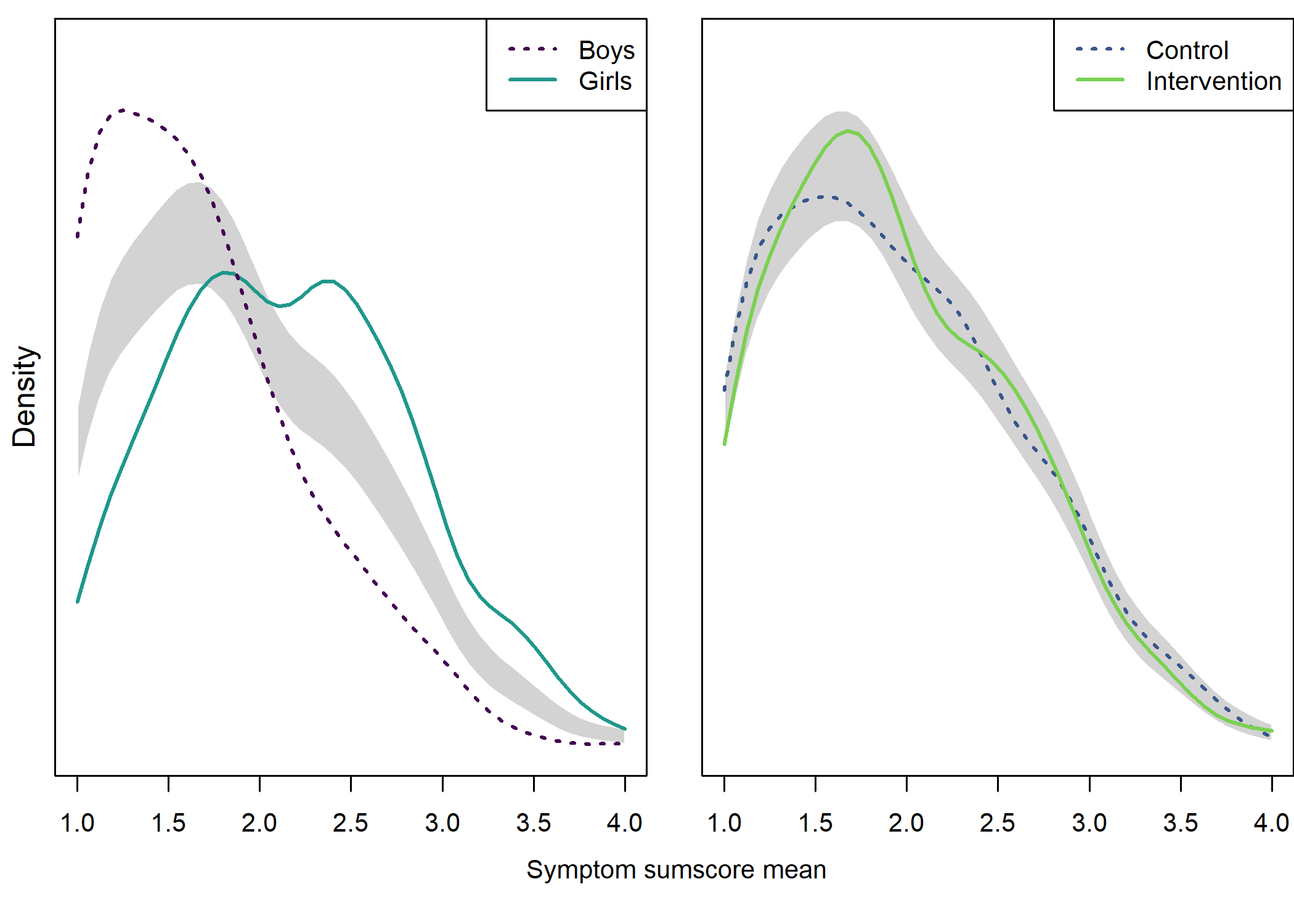
Density plot by intervention and gender
# Create data frame
densplot <- d
levels(densplot$intervention) <- c("Control", "Intervention")
levels(densplot$girl) <- recode(densplot$girl, "boy" = "Boys", "girl" = "Girls")
# This gives side-by-side plots. There's a third plot below the two, which is fake to include x-axis text.
layout(matrix(c(1, 2, 3, 3), nrow = 2, ncol = 2, byrow = TRUE),
widths = c(0.5, 0.5), heights = c(0.45, 0.05))
# Minimise whitespace; see https://stackoverflow.com/questions/15848942/how-to-reduce-space-gap-between-multiple-graphs-in-r
par(mai = c(0.3, 0.3, 0.1, 0.0))
## Girls vs. boys
# Choose only the variables needed and drop NA
dens <- densplot %>% dplyr::select(weartimeAccelerometer_T1, girl) %>%
dplyr::filter(complete.cases(.))
# Set random number generator for reproducibility of bootstrap test of equal densities
set.seed(10)
# Make plot
sm.weartimeAccelerometer_T1_2 <- sm.density.compare2(as.numeric(dens$weartimeAccelerometer_T1),
as.factor(dens$girl),
model = "equal",
xlab = "", ylab = "",
col = viridis::viridis(4, end = 0.8)[c(3, 1)],
lty = c(1,3), yaxt = "n",
bandcol = 'LightGray',
lwd = (c(2,2)))
##
## Test of equal densities: p-value = 0.41
legend("topright", levels(dens$girl), col = viridis::viridis(4, end = 0.8)[c(1, 3)], lty = c(3,1), lwd = (c(2,2)))
mtext(side = 2, "Density", line = 0.5)
## Intervention vs. control
# Choose only the variables needed and drop NA
dens <- densplot %>% dplyr::select(weartimeAccelerometer_T1, intervention) %>%
dplyr::filter(complete.cases(.))
# Set random number generator for reproducibility of bootstrap test of equal densities
set.seed(10)
# Make plot
sm.weartimeAccelerometer_T1_2 <- sm.density.compare2(as.numeric(dens$weartimeAccelerometer_T1),
as.factor(dens$intervention),
model = "equal",
xlab = "", ylab = "",
col = viridis::viridis(4, end = 0.8)[c(2, 4)],
lty = c(3,1), yaxt = "n",
bandcol = 'LightGray',
lwd = (c(2,2)))
##
## Test of equal densities: p-value = 0
legend("topright", levels(dens$intervention), col = viridis::viridis(4, end = 0.8)[c(2, 4)], lty=c(3,1), lwd=(c(2,2)))
# Create x-axis label. See https://stackoverflow.com/questions/11198767/how-to-annotate-across-or-between-plots-in-multi-plot-panels-in-r
par(mar = c(0,0,0,0))
plot(1, 1, type = "n", frame.plot = FALSE, axes = FALSE) # Fake plot for x-axis label
text(x = 1.02, y = 1.3, labels = "Weartime hours", pos = 1)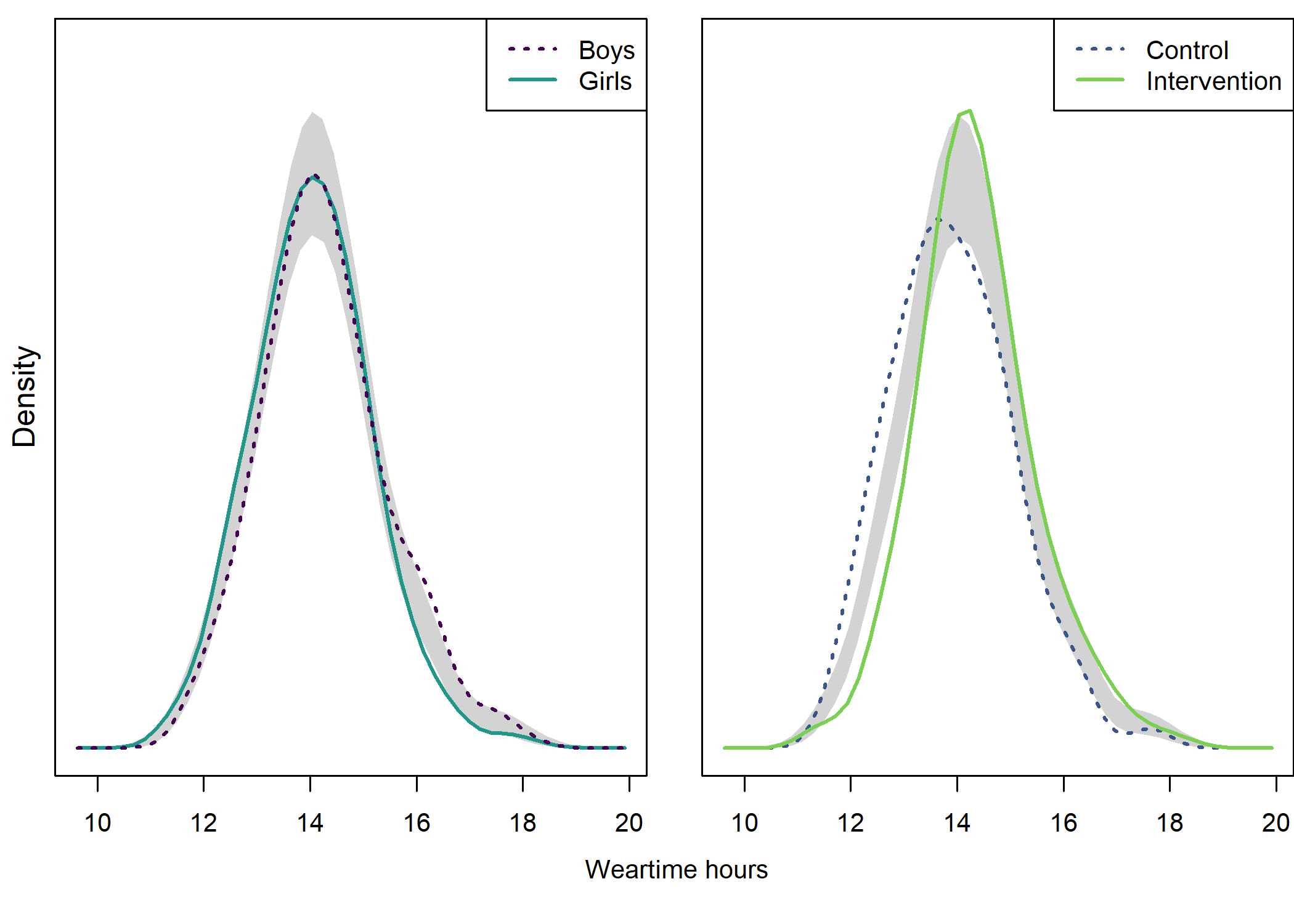
Data preparation
Description
The tab “Information on data preparation” of this section present information on data preparation for the plot
Information on data preparation
Prepare data
# m_weartimeAccelerometer_trackgirl <- brms::bf(weartimeAccelerometer_T1 ~ (1 | track:girl)) %>%
# brms::brm(., data = df, chains = 4, iter = 4000, control = list(adapt_delta = 0.95))
#
# brms::prior_summary(m_weartimeAccelerometer_trackgirl, data = df)
#
# m_weartimeAccelerometer_trackgirl
#
# b_intercept <- brms::fixef(m_weartimeAccelerometer_trackgirl)[1]
#
# # This gives estimates only:
# Weartime_trackgirl_estimates <- brms::ranef(m_weartimeAccelerometer_trackgirl)[[1]][1:10]
#
# # The 2.5%ile:
# Weartime_trackgirl_CIL <- brms::ranef(m_weartimeAccelerometer_trackgirl)[[1]][21:30]
#
# # The 97.5%ile:
# Weartime_trackgirl_CIH <- brms::ranef(m_weartimeAccelerometer_trackgirl)[[1]][31:40]
#
# Weartime_trackgirl_ci <- data.frame(Weartime_trackgirl_CIL, Weartime_trackgirl_estimates, Weartime_trackgirl_CIH, Variable = labels(brms::ranef(m_weartimeAccelerometer_trackgirl))) %>%
# dplyr::mutate(b_intercept = rep(b_intercept, 10)) %>%
# dplyr::mutate(Weartime_trackgirl_CIL = Weartime_trackgirl_CIL + b_intercept,
# Weartime_trackgirl_estimates = Weartime_trackgirl_estimates + b_intercept,
# Weartime_trackgirl_CIH = Weartime_trackgirl_CIH + b_intercept,
# track = c('BA_boy',
# 'BA_girl',
# 'HRC_boy',
# 'HRC_girl',
# 'IT_boy',
# 'IT_girl',
# 'Nur_boy',
# 'Nur_girl',
# 'Other_boy',
# 'Other_girl'
# ))
#
# Weartime_trackgirl_diamonds_girl <- rbind(
# Weartime_trackgirl_ci %>% dplyr::filter(track == "Nur_girl"),
# Weartime_trackgirl_ci %>% dplyr::filter(track == "HRC_girl"),
# Weartime_trackgirl_ci %>% dplyr::filter(track == "BA_girl"),
# Weartime_trackgirl_ci %>% dplyr::filter(track == "IT_girl")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
# Weartime_trackgirl_diamonds_boy <- rbind(
# Weartime_trackgirl_ci %>% dplyr::filter(track == "Nur_boy"),
# Weartime_trackgirl_ci %>% dplyr::filter(track == "HRC_boy"),
# Weartime_trackgirl_ci %>% dplyr::filter(track == "BA_boy"),
# Weartime_trackgirl_ci %>% dplyr::filter(track == "IT_boy")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
#
# m_weartimeAccelerometer_trackintervention <- brms::bf(weartimeAccelerometer_T1 ~ (1 | track:intervention)) %>%
# brms::brm(., data = df, chains = 4, iter = 4000, control = list(adapt_delta = 0.95))
#
# brms::prior_summary(m_weartimeAccelerometer_trackintervention, data = df)
#
# m_weartimeAccelerometer_trackintervention
#
# b_intercept <- brms::fixef(m_weartimeAccelerometer_trackintervention)[1]
#
# # This gives estimates only:
# Weartime_trackintervention_estimates <- brms::ranef(m_weartimeAccelerometer_trackintervention)[[1]][1:10]
#
#
# # The 2.5%ile:
# Weartime_trackintervention_CIL <- brms::ranef(m_weartimeAccelerometer_trackintervention)[[1]][21:30]
#
# # The 97.5%ile:
# Weartime_trackintervention_CIH <- brms::ranef(m_weartimeAccelerometer_trackintervention)[[1]][31:40]
#
# Weartime_trackintervention_ci <- data.frame(Weartime_trackintervention_CIL, Weartime_trackintervention_estimates, Weartime_trackintervention_CIH, Variable = labels(brms::ranef(m_weartimeAccelerometer_trackintervention))) %>%
# dplyr::mutate(b_intercept = rep(b_intercept, 10)) %>%
# dplyr::mutate(Weartime_trackintervention_CIL = Weartime_trackintervention_CIL + b_intercept,
# Weartime_trackintervention_estimates = Weartime_trackintervention_estimates + b_intercept,
# Weartime_trackintervention_CIH = Weartime_trackintervention_CIH + b_intercept,
# track = c('BA_control',
# 'BA_intervention',
# 'HRC_control',
# 'HRC_intervention',
# 'IT_control',
# 'IT_intervention',
# 'Nur_control',
# 'Nur_intervention',
# 'Other_control',
# 'Other_intervention'
# ))
#
# Weartime_trackintervention_diamonds_intervention <- rbind(
# Weartime_trackintervention_ci %>% dplyr::filter(track == "Nur_intervention"),
# Weartime_trackintervention_ci %>% dplyr::filter(track == "HRC_intervention"),
# Weartime_trackintervention_ci %>% dplyr::filter(track == "BA_intervention"),
# Weartime_trackintervention_ci %>% dplyr::filter(track == "IT_intervention")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
# Weartime_trackintervention_diamonds_control <- rbind(
# Weartime_trackintervention_ci %>% dplyr::filter(track == "Nur_control"),
# Weartime_trackintervention_ci %>% dplyr::filter(track == "HRC_control"),
# Weartime_trackintervention_ci %>% dplyr::filter(track == "BA_control"),
# Weartime_trackintervention_ci %>% dplyr::filter(track == "IT_control")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
# save(Weartime_trackgirl_diamonds_girl, file = "./Rdata_files/Weartime_trackgirl_diamonds_girl.Rdata")
# save(Weartime_trackgirl_diamonds_boy, file = "./Rdata_files/Weartime_trackgirl_diamonds_boy.Rdata")
# save(Weartime_trackintervention_diamonds_intervention, file = "./Rdata_files/Weartime_trackintervention_diamonds_intervention.Rdata")
# save(Weartime_trackintervention_diamonds_control, file = "./Rdata_files/Weartime_trackintervention_diamonds_control.Rdata")
load("./Rdata_files/Weartime_trackgirl_diamonds_girl.Rdata")
load("./Rdata_files/Weartime_trackgirl_diamonds_boy.Rdata")
load("./Rdata_files/Weartime_trackintervention_diamonds_intervention.Rdata")
load("./Rdata_files/Weartime_trackintervention_diamonds_control.Rdata")
Weartime_trackgirl_diamonds_girlSensitivity analyses and robustness checks
Ideally, one would perform sensitivity analyses and robustness checks; e.g. comparing the estimates with frequentist multilevel models and following the WAMBS-checklist. Due to resource constraints, we are forced to forego this phase of analysis and instead, only show the linear model results for intercepts in each intervention-gender-track combination separately.
# To test the code with a single variable:
df_for_models_nested <- df %>%
dplyr::select(track, weartimeAccelerometer_T1, girl, intervention) %>%
dplyr::mutate(track = forcats::fct_recode(track, NULL = "Other")) %>%
na.omit() %>%
dplyr::group_by(girl, intervention, track) %>%
tidyr::nest()
# This produced a data frame with columns "girl", "intervention", "track" and "data", the last of which is a data frame in each cell. E.g. for row wit intervention group girls in IT, there's a data frame for their values in the last column.
df_fitted <- df_for_models_nested %>%
dplyr::mutate(fit = map(data, ~ lm(weartimeAccelerometer_T1 ~ 1, data = .x)))
# Now there's a linear model for each combination in the data frame
df_fitted <- df_fitted %>%
dplyr::mutate(
mean = map_dbl(fit, ~ coef(.x)[["(Intercept)"]]),
ci_low = map_dbl(fit, ~ confint(.x)["(Intercept)", 1]),
ci_high = map_dbl(fit, ~ confint(.x)["(Intercept)", 2]),
nonmissings = map_dbl(fit, ~ nobs(.x))
) %>%
dplyr::mutate(mean = round(mean, 2),
ci_low = round(ci_low, 2),
ci_high = round(ci_high, 2))
## (here was an earlier attempt for random effects)
# df_fitted <- df_fitted %>%
# dplyr::mutate(
# mean = map_dbl(fit, ~ lme4::fixef(.x)),
# m_p = map(fit, ~ profile(.x)),
# ci_low = map_dbl(m_p, ~ confint(.x)["(Intercept)", 1]),
# ci_high = map_dbl(m_p, ~ confint(.x)["(Intercept)", 2]),
# nonmissings = map_dbl(fit, ~ length(.x@resp$y))
# )
## (here was was an earlier attempt to use sandwich estimator
# df_for_sandwich_nested_withClusters <- df_for_sandwich_nested %>%
# dplyr::mutate(sandwich_clusters = purrr::map(data, magrittr::extract, c("group", "school", "track")))
# The last line selected the cluster variables from the data frame of each intervention-gender combination)
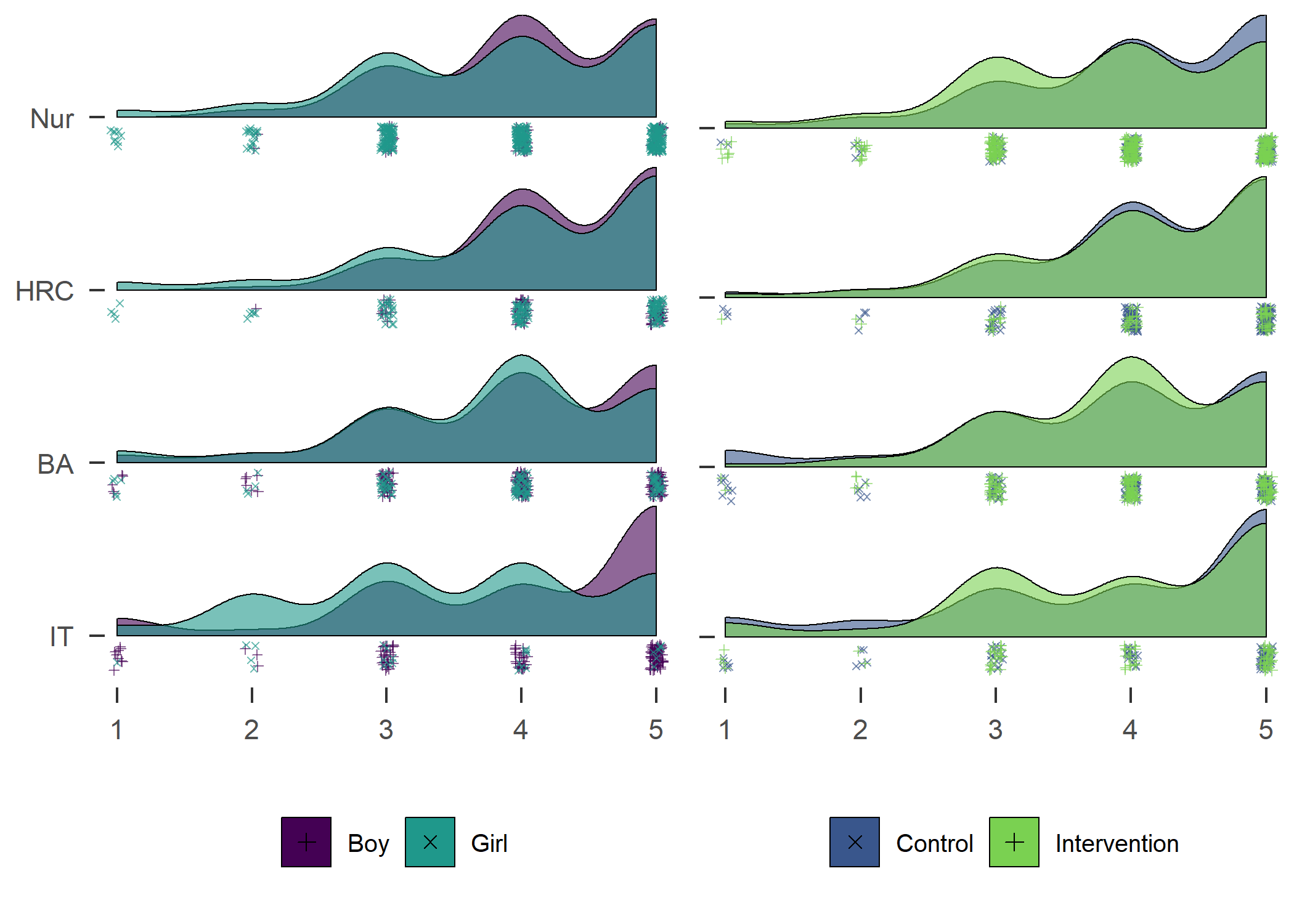
DT::datatable(df_fitted)Density plot of girls and boys in different educational tracks and schools
df %>% dplyr::select(weartimeAccelerometer_T1, girl, trackSchool) %>%
group_by(girl, trackSchool) %>%
summarise(mean = mean(weartimeAccelerometer_T1, na.rm = TRUE),
median = median(weartimeAccelerometer_T1, na.rm = TRUE),
max = max(weartimeAccelerometer_T1, na.rm = TRUE),
min = min(weartimeAccelerometer_T1, na.rm = TRUE),
sd = sd(weartimeAccelerometer_T1, na.rm = TRUE),
n = n()) %>%
DT::datatable() #papaja::apa_table()
df %>% dplyr::select(weartimeAccelerometer_T1, girl, trackSchool) %>%
group_by(girl) %>%
summarise(mean = mean(weartimeAccelerometer_T1, na.rm = TRUE),
median = median(weartimeAccelerometer_T1, na.rm = TRUE),
max = max(weartimeAccelerometer_T1, na.rm = TRUE),
min = min(weartimeAccelerometer_T1, na.rm = TRUE),
sd = sd(weartimeAccelerometer_T1, na.rm = TRUE),
n = n()) %>%
DT::datatable() #papaja::apa_table()
plot1 <- df %>% dplyr::select(id,
trackSchool = trackSchool,
girl,
Weartime = weartimeAccelerometer_T1) %>%
dplyr::filter(!is.na(trackSchool), !grepl('Other|NANA|BA4|HRC2|Nur2', trackSchool)) %>% # Drop categories with just few participants
ggplot2::ggplot(aes(y = trackSchool)) +
ggridges::geom_density_ridges2(aes(x = Weartime, colour = "black",
fill = paste(trackSchool, girl)),
scale = 1,
alpha = 0.6, size = 0.25,
from = 0, #to = 450,
jittered_points=TRUE, point_shape=21,
point_fill="black") +
labs(x = "",
y = "") +
scale_y_discrete(expand = c(0.01, 0)) +
scale_x_continuous(expand = c(0.01, 0)) +
ggridges::scale_fill_cyclical(
labels = c('BA1 boy' = "Boy", 'BA1 girl' = "Girl"),
values = viridis::viridis(4, end = 0.8)[c(1, 3)],
name = "", guide = guide_legend(override.aes = list(alpha = 1))) +
ggridges::scale_colour_cyclical(values = "black") +
ggridges::theme_ridges(grid = FALSE) +
theme(legend.position="bottom", axis.text=element_text(size=10))
plot1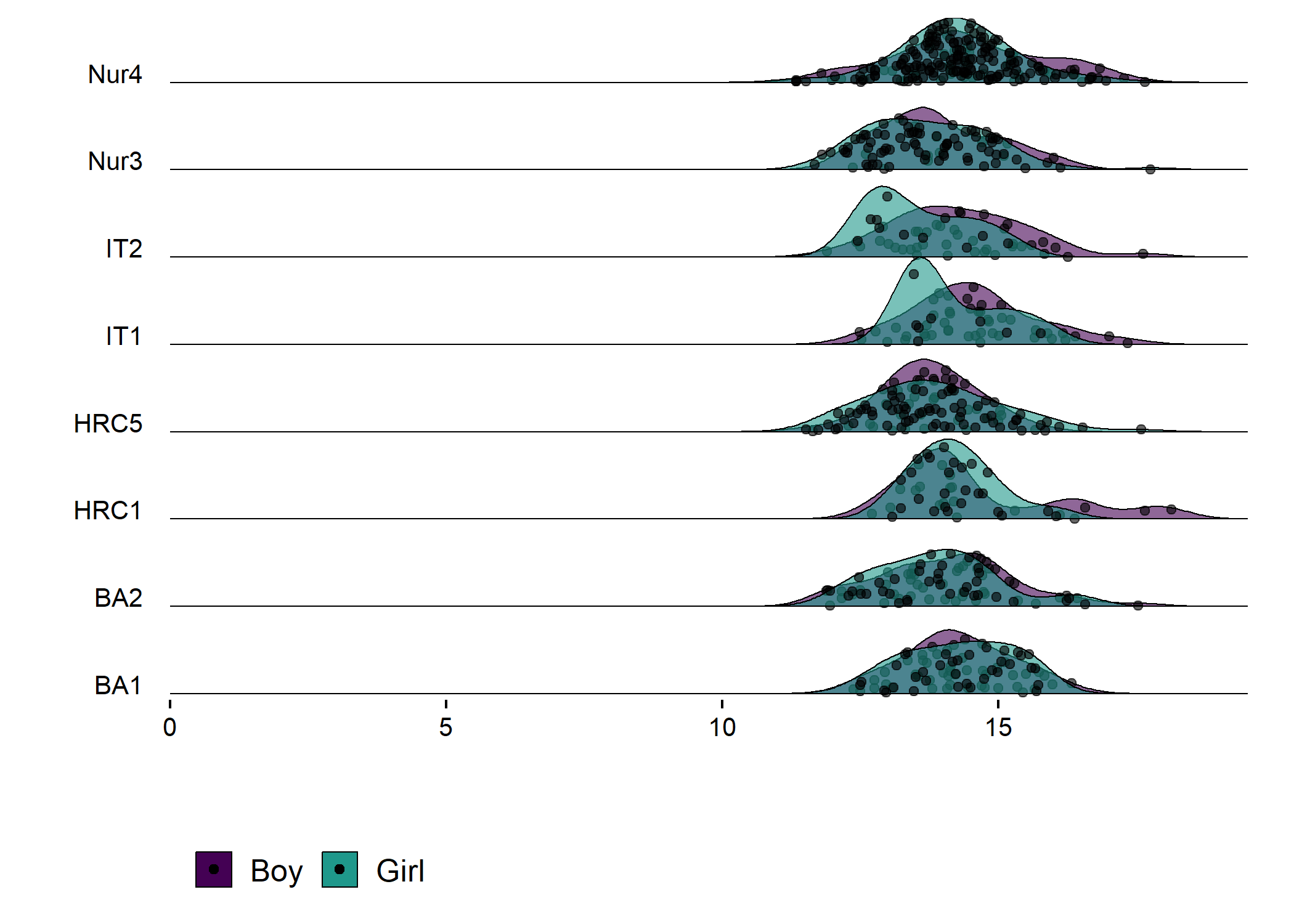
Density plot by track
plot1 <- df %>% dplyr::select(id,
track = track,
girl,
Weartime = weartimeAccelerometer_T1) %>%
dplyr::filter(!is.na(track), track != "Other") %>% # Drop category "Other"
dplyr::mutate(track = factor(track, levels = c("IT", "BA", "HRC", "Nur")),
track = recode_factor(track, "IT" = "IT", "BA" = "BA",
"HRC" = "HRC", "Nur" = "Nur")) %>%
ggplot2::ggplot(aes(y = track)) +
ggridges::geom_density_ridges2(aes(x = Weartime, colour = "black",
fill = paste(track, girl),
point_color = girl,
point_fill = girl,
point_shape = girl),
scale = .75, alpha = 0.6, size = 0.25,
position = position_raincloud(width = 0.05, height = 0.15),
# from = 0, to = 450,
jittered_points = TRUE,
point_size = 1) +
labs(x = NULL,
y = NULL) +
scale_y_discrete(expand = c(0.1, 0)) +
scale_x_continuous(expand = c(0.01, 0)) +
ggridges::scale_fill_cyclical(
labels = c('BA boy' = "Boy", 'BA girl' = "Girl"),
values = viridis::viridis(4, end = 0.8)[c(1, 3)],
name = "",
guide = guide_legend(override.aes = list(alpha = 1,
point_shape = c(3, 4),
point_size = 2))) +
ggridges::scale_colour_cyclical(values = "black") +
ggridges::theme_ridges(grid = FALSE) +
# ggplot2::scale_fill_manual(values = c("#A0FFA0", "#A0A0FF")) +
ggridges::scale_discrete_manual(aesthetics = "point_color",
values = viridis::viridis(4, end = 0.8)[c(3, 1)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_fill",
values = viridis::viridis(4, end = 0.8)[c(3, 1)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_shape",
values = c(4, 3),
guide = "none") +
papaja::theme_apa() +
theme(legend.position = "bottom")
Weartime_trackgirl_diamonds_girl_hours <- Weartime_trackgirl_diamonds_girl %>%
dplyr::mutate_if(is.numeric, ~./60)
Weartime_trackgirl_diamonds_boy_hours <- Weartime_trackgirl_diamonds_boy %>%
dplyr::mutate_if(is.numeric, ~./60)
plot1 <- plot1 +
userfriendlyscience::diamondPlot(Weartime_trackgirl_diamonds_girl_hours,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[3],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15)) + # specify y-position; option to move the diamond
userfriendlyscience::diamondPlot(Weartime_trackgirl_diamonds_boy_hours,
returnLayerOnly = TRUE,
lineColor = "black",
linetype = "solid",
color=viridis::viridis(4, end = 0.8)[1],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15))
# Draw plot with intervention and control densities
plot2 <- df %>% dplyr::select(id,
track = track,
intervention,
Weartime = weartimeAccelerometer_T1) %>%
dplyr::filter(!is.na(track), track != "Other") %>% # Drop category "Other"
dplyr::mutate(track = factor(track, levels = c("IT", "BA", "HRC", "Nur")),
track = recode_factor(track, "IT" = "IT", "BA" = "BA",
"HRC" = "HRC", "Nur" = "Nur")) %>%
ggplot2::ggplot(aes(y = track)) +
ggridges::geom_density_ridges2(aes(x = Weartime, colour = "black",
fill = paste(track, intervention),
point_color = intervention,
point_fill = intervention,
point_shape = intervention),
scale = .75, alpha = 0.6, size = 0.25,
position = position_raincloud(width = 0.05, height = 0.15),
# from = 0, to = 450,
jittered_points = TRUE,
point_size = 1) +
labs(x = NULL,
y = NULL) +
scale_y_discrete(expand = c(0.1, 0), labels = NULL) +
scale_x_continuous(expand = c(0.01, 0)) +
ggridges::scale_fill_cyclical(
labels = c('BA 0' = "Control", 'BA 1' = "Intervention"),
values = viridis::viridis(4, end = 0.8)[c(2, 4)],
name = "",
guide = guide_legend(override.aes = list(alpha = 1,
point_shape = c(4, 3),
point_size = 2))) +
ggridges::scale_colour_cyclical(values = "black") +
ggridges::theme_ridges(grid = FALSE) +
# ggplot2::scale_fill_manual(values = c("#A0FFA0", "#A0A0FF")) +
ggridges::scale_discrete_manual(aesthetics = "point_color",
values = viridis::viridis(4, end = 0.8)[c(2, 4)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_fill",
values = viridis::viridis(4, end = 0.8)[c(2, 4)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_shape",
values = c(4, 3),
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_alpha",
values = 0.06,
guide = "none") +
papaja::theme_apa() +
theme(legend.position = "bottom")
Weartime_trackintervention_diamonds_intervention_hours <- Weartime_trackintervention_diamonds_intervention %>%
dplyr::mutate_if(is.numeric, ~./60)
Weartime_trackintervention_diamonds_control_hours <- Weartime_trackintervention_diamonds_control %>%
dplyr::mutate_if(is.numeric, ~./60)
plot2 <- plot2 +
userfriendlyscience::diamondPlot(Weartime_trackintervention_diamonds_intervention_hours,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[4],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15)) +
userfriendlyscience::diamondPlot(Weartime_trackintervention_diamonds_control_hours,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[2],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15))
plot1 + plot2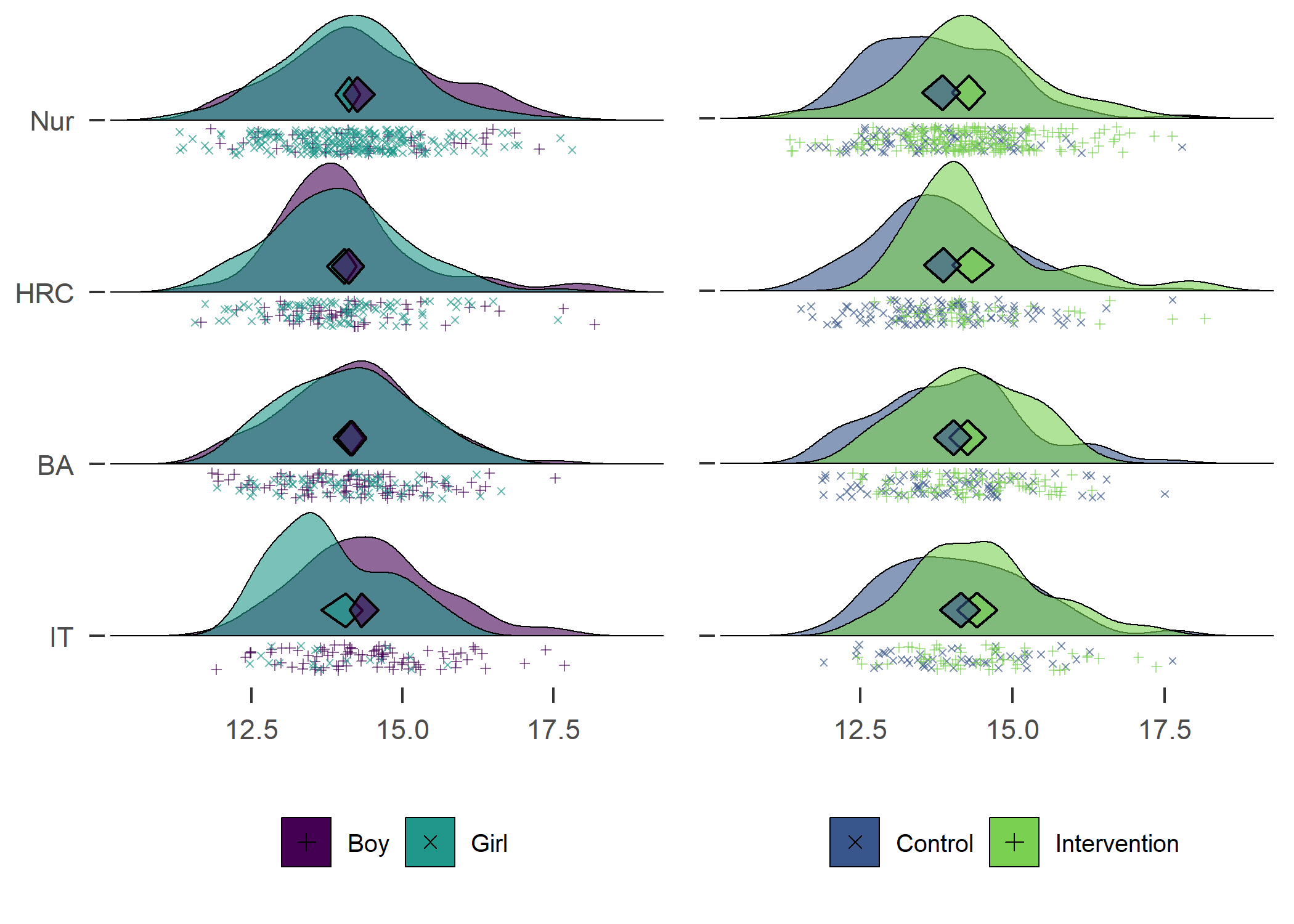
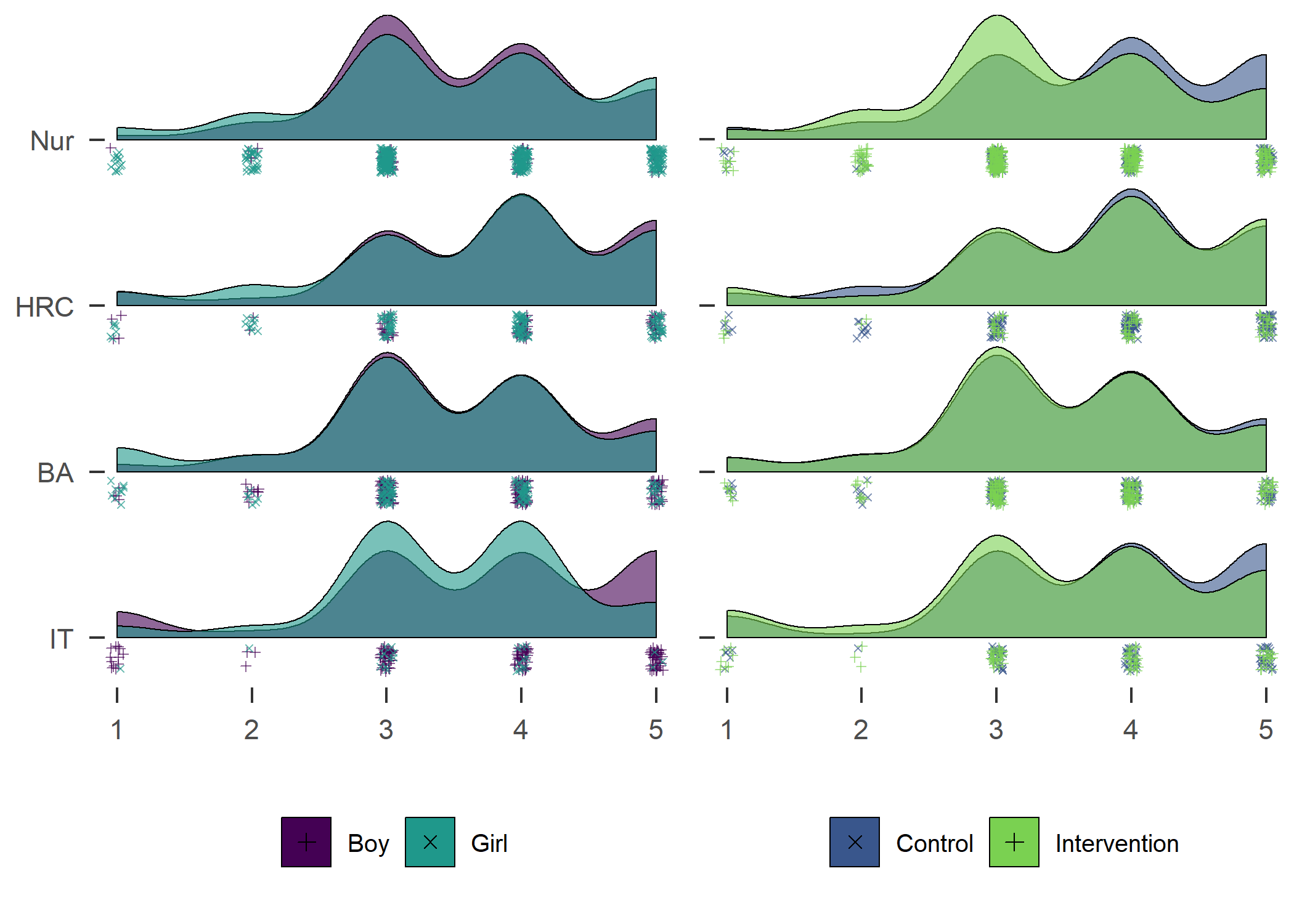
Accelerometer wear time. Nur = Practical nurse, HRC = Hotel, restaurant and catering, BA = Business and administration, IT = Information and communications technology.
Accelerometer-measured MVPA
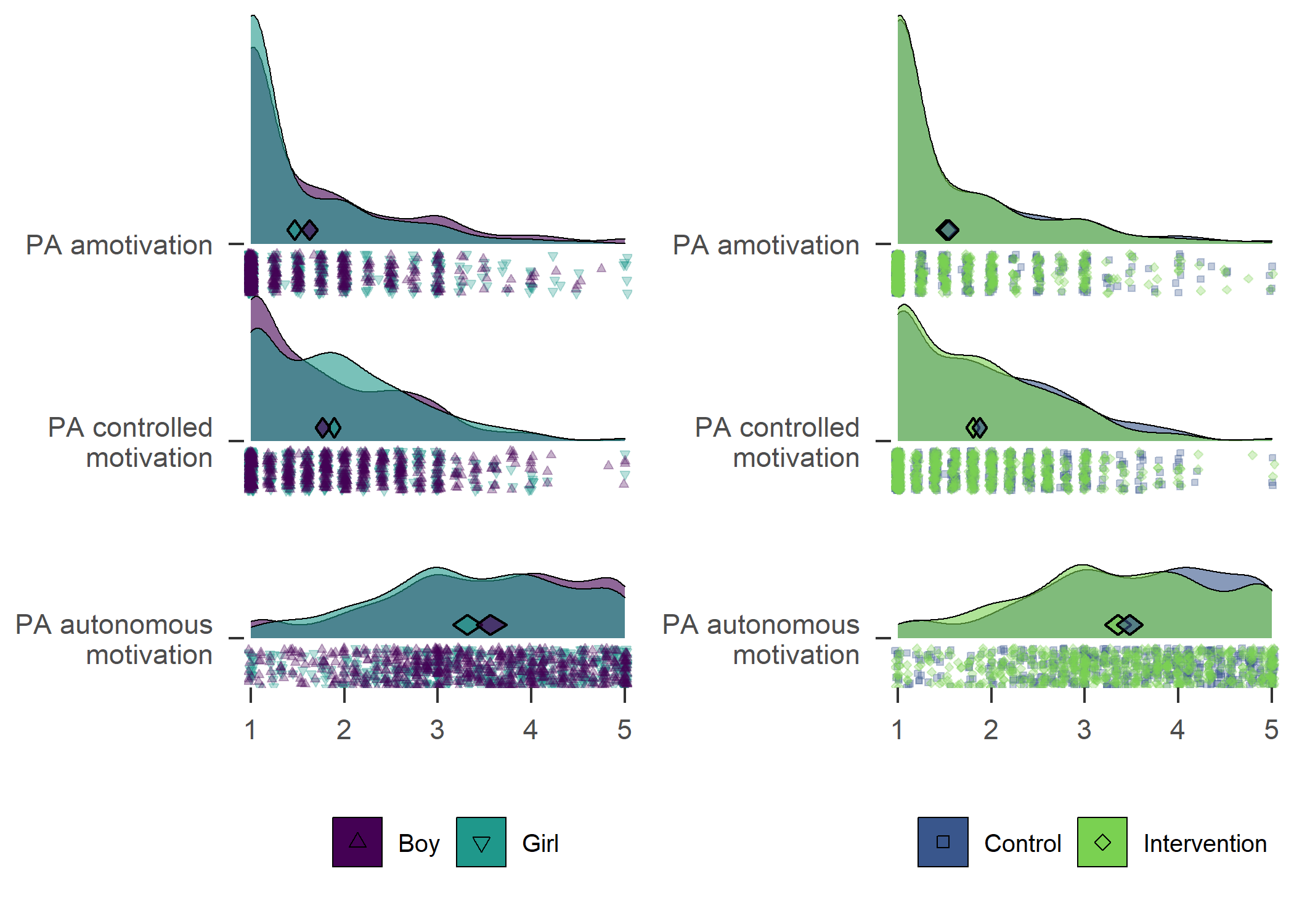
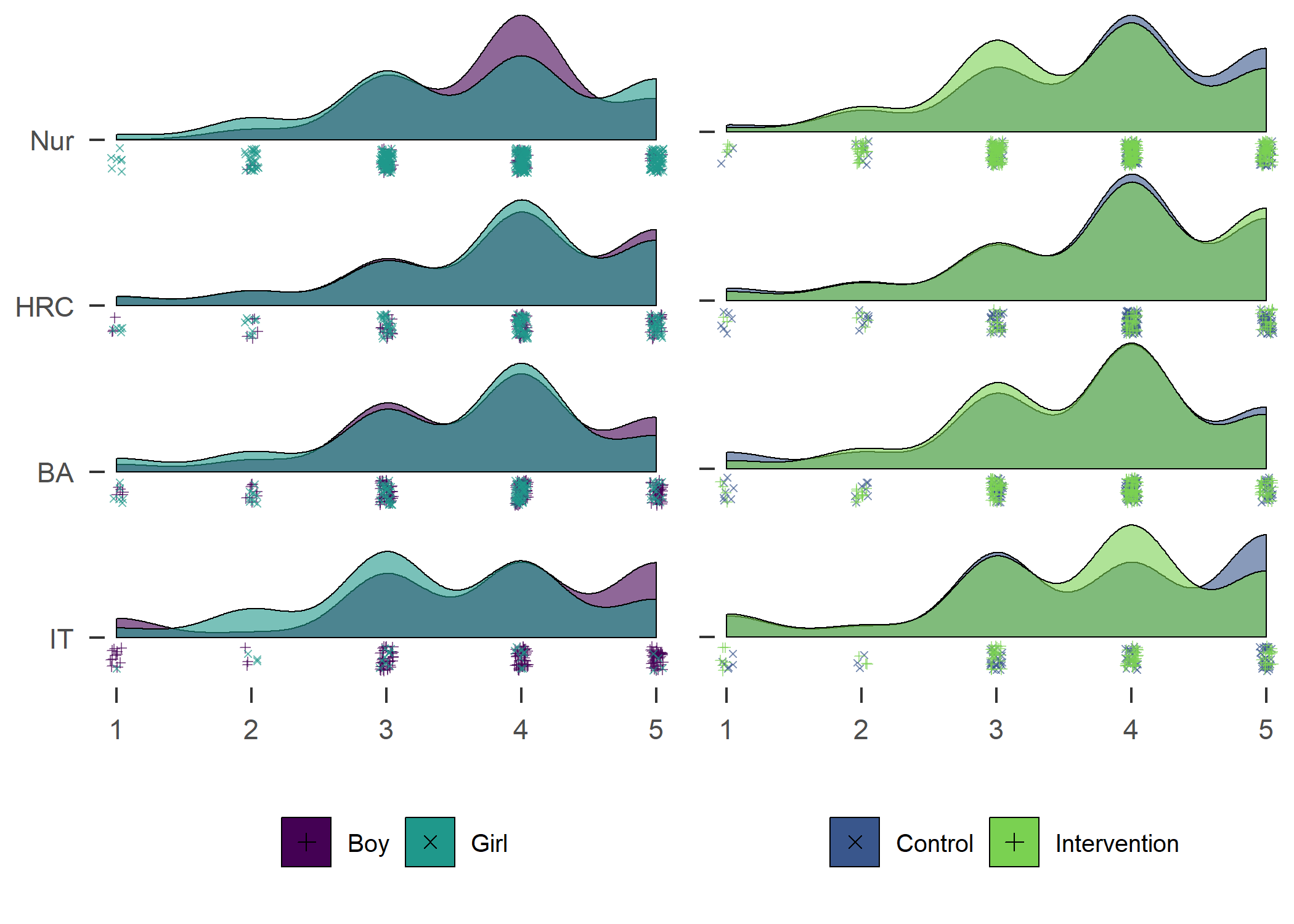
Contrary to some other findings, in our sample girls were more active than boys.
MVPAgirl_df <- df %>% group_by(girl) %>%
dplyr::select(girl, mvpaAccelerometer_T1) %>%
summarise(`mean accelerometer-measured MVPA difference between girls and boys` = mean(mvpaAccelerometer_T1, na.rm = TRUE),
`median accelerometer-measured MVPA difference between girls and boys` = median(mvpaAccelerometer_T1, na.rm = TRUE))
userfriendlyscience::meanDiff(df$girl, df$mvpaAccelerometer_T1)
## Input variables:
##
## girl (grouping variable)
## mvpaAccelerometer_T1 (dependent variable)
## Mean 1 (girl) = 1.08, sd = 0.47, n = 437
## Mean 2 (boy)= 1.11, sd = 0.54, n = 294
##
## Independent samples t-test (tested for equal variances, p = .006, so unequal variances)
## (standard deviation used of largest sample, 0.47)
##
## 95% confidence intervals:
## Absolute mean difference: [-0.11, 0.04] (Absolute mean difference: -0.03)
## Cohen's d for difference: [-0.22, 0.08] (Cohen's d point estimate: -0.07)
## Hedges g for difference: [-0.22, 0.08] (Hedges g point estimate: -0.07)
##
## Achieved power for d=-0.07: 0.1612 (for small: 0.7704; medium: 1; large: 1)
##
## (secondary information (NHST): t[565.85] = -0.86, p = .389)
##
##
## NOTE: because the t-test is based on unequal variances, the NHST p-value may be inconsistent with the confidence interval. Although this is not a problem, if you wish to ensure consistency, you can use parameter "var.equal = 'yes'" to force equal variances.Density plot by intervention and gender
# Create data frame
densplot <- d
levels(densplot$intervention) <- c("Control", "Intervention")
levels(densplot$girl) <- recode(densplot$girl, "boy" = "Boys", "girl" = "Girls")
# This gives side-by-side plots. There's a third plot below the two, which is fake to include x-axis text.
layout(matrix(c(1, 2, 3, 3), nrow = 2, ncol = 2, byrow = TRUE),
widths = c(0.5, 0.5), heights = c(0.45, 0.05))
# Minimise whitespace; see https://stackoverflow.com/questions/15848942/how-to-reduce-space-gap-between-multiple-graphs-in-r
par(mai = c(0.3, 0.3, 0.1, 0.0))
## Girls vs. boys
# Choose only the variables needed and drop NA
dens <- densplot %>% dplyr::select(mvpaAccelerometer_T1, girl) %>%
dplyr::filter(complete.cases(.))
# Set random number generator for reproducibility of bootstrap test of equal densities
set.seed(10)
# Make plot
sm.mvpaAccelerometer_T1_2 <- sm.density.compare2(as.numeric(dens$mvpaAccelerometer_T1),
as.factor(dens$girl),
model = "equal",
xlab = "", ylab = "",
col = viridis::viridis(4, end = 0.8)[c(3, 1)],
lty = c(1,3), yaxt = "n",
bandcol = 'LightGray',
lwd = (c(2,2)))
##
## Test of equal densities: p-value = 0.12
legend("topright", levels(dens$girl), col = viridis::viridis(4, end = 0.8)[c(1, 3)], lty = c(3,1), lwd = (c(2,2)))
mtext(side = 2, "Density", line = 0.5)
## Intervention vs. control
# Choose only the variables needed and drop NA
dens <- densplot %>% dplyr::select(mvpaAccelerometer_T1, intervention) %>%
dplyr::filter(complete.cases(.))
# Set random number generator for reproducibility of bootstrap test of equal densities
set.seed(10)
# Make plot
sm.mvpaAccelerometer_T1_2 <- sm.density.compare2(as.numeric(dens$mvpaAccelerometer_T1),
as.factor(dens$intervention),
model = "equal",
xlab = "", ylab = "",
col = viridis::viridis(4, end = 0.8)[c(2, 4)],
lty = c(3,1), yaxt = "n",
bandcol = 'LightGray',
lwd = (c(2,2)))
##
## Test of equal densities: p-value = 0.14
legend("topright", levels(dens$intervention), col = viridis::viridis(4, end = 0.8)[c(2, 4)], lty=c(3,1), lwd=(c(2,2)))
# Create x-axis label. See https://stackoverflow.com/questions/11198767/how-to-annotate-across-or-between-plots-in-multi-plot-panels-in-r
par(mar = c(0,0,0,0))
plot(1, 1, type = "n", frame.plot = FALSE, axes = FALSE) # Fake plot for x-axis label
text(x = 1.02, y = 1.3, labels = "MVPA hours", pos = 1)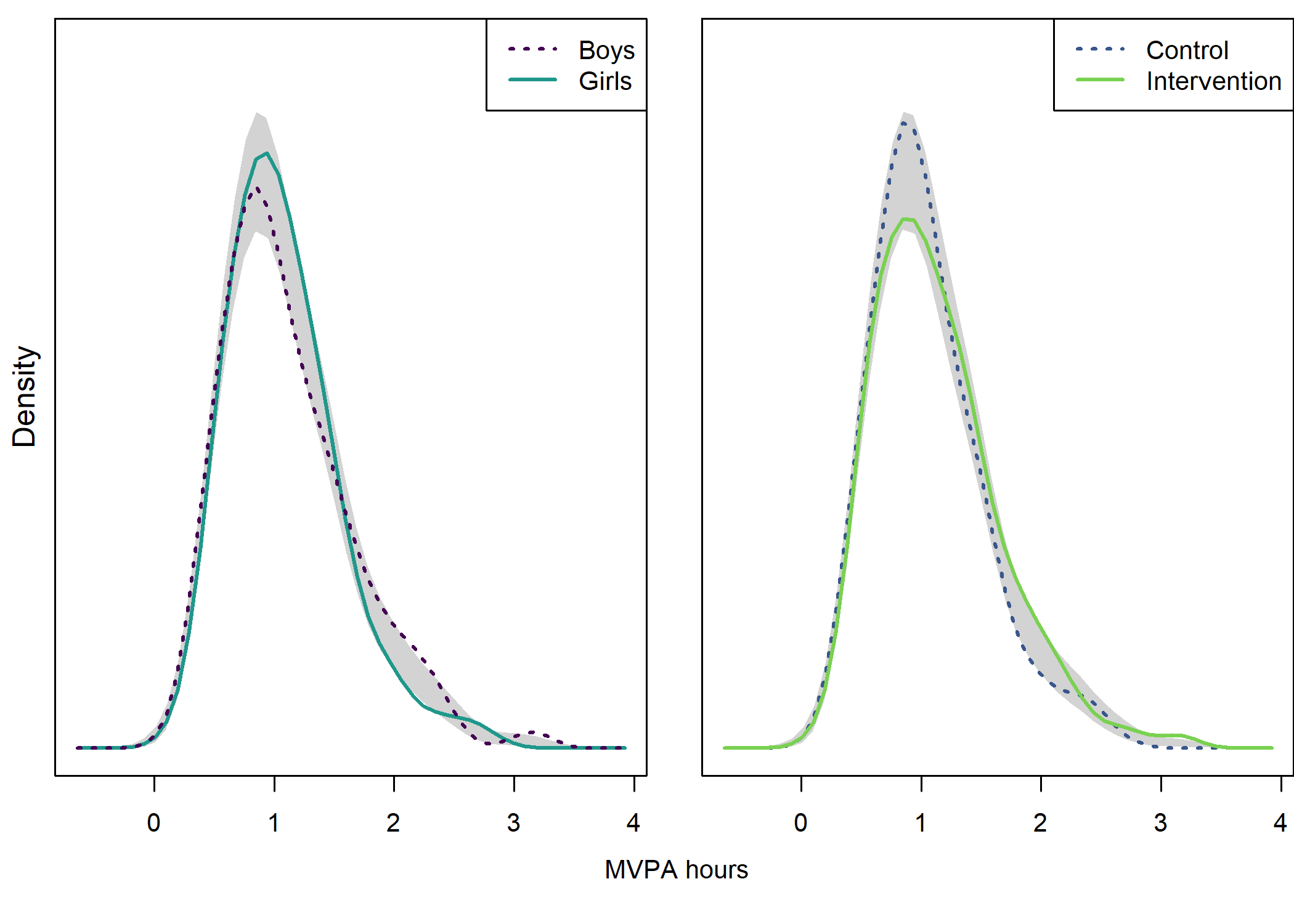
Data preparation
Description
The tab “Information on data preparation” of this section present information on data preparation for the plot \(~\) \(~\)
Information on data preparation
Prepare data
# m_mvpaAccelerometer_trackgirl <- brms::bf(mvpaAccelerometer_T1 ~ (1 | track:girl)) %>%
# brms::brm(., data = df, chains = 4, iter = 4000, control = list(adapt_delta = 0.95))
#
# brms::prior_summary(m_mvpaAccelerometer_trackgirl, data = df)
#
# m_mvpaAccelerometer_trackgirl
#
# b_intercept <- brms::fixef(m_mvpaAccelerometer_trackgirl)[1]
#
# # This gives estimates only:
# PA_trackgirl_estimates <- brms::ranef(m_mvpaAccelerometer_trackgirl)[[1]][1:10]
#
# # The 2.5%ile:
# PA_trackgirl_CIL <- brms::ranef(m_mvpaAccelerometer_trackgirl)[[1]][21:30]
#
# # The 97.5%ile:
# PA_trackgirl_CIH <- brms::ranef(m_mvpaAccelerometer_trackgirl)[[1]][31:40]
#
# PA_trackgirl_ci <- data.frame(PA_trackgirl_CIL, PA_trackgirl_estimates, PA_trackgirl_CIH, Variable = labels(brms::ranef(m_mvpaAccelerometer_trackgirl))) %>%
# dplyr::mutate(b_intercept = rep(b_intercept, 10)) %>%
# dplyr::mutate(PA_trackgirl_CIL = PA_trackgirl_CIL + b_intercept,
# PA_trackgirl_estimates = PA_trackgirl_estimates + b_intercept,
# PA_trackgirl_CIH = PA_trackgirl_CIH + b_intercept,
# track = c('BA_boy',
# 'BA_girl',
# 'HRC_boy',
# 'HRC_girl',
# 'IT_boy',
# 'IT_girl',
# 'Nur_boy',
# 'Nur_girl',
# 'Other_boy',
# 'Other_girl'
# ))
#
# PA_trackgirl_diamonds_girl <- rbind(
# PA_trackgirl_ci %>% dplyr::filter(track == "Nur_girl"),
# PA_trackgirl_ci %>% dplyr::filter(track == "HRC_girl"),
# PA_trackgirl_ci %>% dplyr::filter(track == "BA_girl"),
# PA_trackgirl_ci %>% dplyr::filter(track == "IT_girl")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
# PA_trackgirl_diamonds_boy <- rbind(
# PA_trackgirl_ci %>% dplyr::filter(track == "Nur_boy"),
# PA_trackgirl_ci %>% dplyr::filter(track == "HRC_boy"),
# PA_trackgirl_ci %>% dplyr::filter(track == "BA_boy"),
# PA_trackgirl_ci %>% dplyr::filter(track == "IT_boy")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
#
# m_mvpaAccelerometer_trackintervention <- brms::bf(mvpaAccelerometer_T1 ~ (1 | track:intervention)) %>%
# brms::brm(., data = df, chains = 4, iter = 4000, control = list(adapt_delta = 0.95))
#
# brms::prior_summary(m_mvpaAccelerometer_trackintervention, data = df)
#
# m_mvpaAccelerometer_trackintervention
#
# b_intercept <- brms::fixef(m_mvpaAccelerometer_trackintervention)[1]
#
# # This gives estimates only:
# PA_trackintervention_estimates <- brms::ranef(m_mvpaAccelerometer_trackintervention)[[1]][1:10]
#
#
# # The 2.5%ile:
# PA_trackintervention_CIL <- brms::ranef(m_mvpaAccelerometer_trackintervention)[[1]][21:30]
#
# # The 97.5%ile:
# PA_trackintervention_CIH <- brms::ranef(m_mvpaAccelerometer_trackintervention)[[1]][31:40]
#
# PA_trackintervention_ci <- data.frame(PA_trackintervention_CIL, PA_trackintervention_estimates, PA_trackintervention_CIH, Variable = labels(brms::ranef(m_mvpaAccelerometer_trackintervention))) %>%
# dplyr::mutate(b_intercept = rep(b_intercept, 10)) %>%
# dplyr::mutate(PA_trackintervention_CIL = PA_trackintervention_CIL + b_intercept,
# PA_trackintervention_estimates = PA_trackintervention_estimates + b_intercept,
# PA_trackintervention_CIH = PA_trackintervention_CIH + b_intercept,
# track = c('BA_control',
# 'BA_intervention',
# 'HRC_control',
# 'HRC_intervention',
# 'IT_control',
# 'IT_intervention',
# 'Nur_control',
# 'Nur_intervention',
# 'Other_control',
# 'Other_intervention'
# ))
#
# PA_trackintervention_diamonds_intervention <- rbind(
# PA_trackintervention_ci %>% dplyr::filter(track == "Nur_intervention"),
# PA_trackintervention_ci %>% dplyr::filter(track == "HRC_intervention"),
# PA_trackintervention_ci %>% dplyr::filter(track == "BA_intervention"),
# PA_trackintervention_ci %>% dplyr::filter(track == "IT_intervention")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
# PA_trackintervention_diamonds_control <- rbind(
# PA_trackintervention_ci %>% dplyr::filter(track == "Nur_control"),
# PA_trackintervention_ci %>% dplyr::filter(track == "HRC_control"),
# PA_trackintervention_ci %>% dplyr::filter(track == "BA_control"),
# PA_trackintervention_ci %>% dplyr::filter(track == "IT_control")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
# # readr::write_rds(PA_trackgirl_diamonds_girl,
# # path = "./Rdata_files/PA_trackgirl_diamonds_girl.RDS")
# # readr::write_rds(PA_trackgirl_diamonds_boy,
# # path = "./Rdata_files/PA_trackgirl_diamonds_boy.RDS")
# # readr::write_rds(PA_trackintervention_diamonds_intervention,
# # path = "./Rdata_files/PA_trackintervention_diamonds_intervention.RDS")
# # readr::write_rds(PA_trackintervention_diamonds_control,
# # path = "./Rdata_files/PA_trackintervention_diamonds_control.RDS")
#
# save(PA_trackgirl_diamonds_girl, file = "./Rdata_files/PA_trackgirl_diamonds_girl.Rdata")
# save(PA_trackgirl_diamonds_boy, file = "./Rdata_files/PA_trackgirl_diamonds_boy.Rdata")
# save(PA_trackintervention_diamonds_intervention, file = "./Rdata_files/PA_trackintervention_diamonds_intervention.Rdata")
# save(PA_trackintervention_diamonds_control, file = "./Rdata_files/PA_trackintervention_diamonds_control.Rdata")
load("./Rdata_files/PA_trackgirl_diamonds_girl.Rdata")
load("./Rdata_files/PA_trackgirl_diamonds_boy.Rdata")
load("./Rdata_files/PA_trackintervention_diamonds_intervention.Rdata")
load("./Rdata_files/PA_trackintervention_diamonds_control.Rdata")
PA_trackgirl_diamonds_girlSensitivity analyses and robustness checks
Ideally, one would perform sensitivity analyses and robustness checks; e.g. comparing the estimates with frequentist multilevel models and following the WAMBS-checklist. Due to resource constraints, we are forced to forego this phase of analysis and instead, only show the linear model results for intercepts in each intervention-gender-track combination separately.
# To test the code with a single variable:
df_for_models_nested <- df %>%
dplyr::select(track, mvpaAccelerometer_T1, girl, intervention) %>%
dplyr::mutate(track = forcats::fct_recode(track, NULL = "Other")) %>%
na.omit() %>%
dplyr::group_by(girl, intervention, track) %>%
tidyr::nest()
# This produced a data frame with columns "girl", "intervention", "track" and "data", the last of which is a data frame in each cell. E.g. for row wit intervention group girls in IT, there's a data frame for their values in the last column.
df_fitted <- df_for_models_nested %>%
dplyr::mutate(fit = map(data, ~ lm(mvpaAccelerometer_T1 ~ 1, data = .x)))
# Now there's a linear model for each combination in the data frame
df_fitted <- df_fitted %>%
dplyr::mutate(
mean = map_dbl(fit, ~ coef(.x)[["(Intercept)"]]),
ci_low = map_dbl(fit, ~ confint(.x)["(Intercept)", 1]),
ci_high = map_dbl(fit, ~ confint(.x)["(Intercept)", 2]),
nonmissings = map_dbl(fit, ~ nobs(.x))
) %>%
dplyr::mutate(mean = round(mean, 2),
ci_low = round(ci_low, 2),
ci_high = round(ci_high, 2))
## (here was an earlier attempt for random effects)
# df_fitted <- df_fitted %>%
# dplyr::mutate(
# mean = map_dbl(fit, ~ lme4::fixef(.x)),
# m_p = map(fit, ~ profile(.x)),
# ci_low = map_dbl(m_p, ~ confint(.x)["(Intercept)", 1]),
# ci_high = map_dbl(m_p, ~ confint(.x)["(Intercept)", 2]),
# nonmissings = map_dbl(fit, ~ length(.x@resp$y))
# )
## (here was was an earlier attempt to use sandwich estimator
# df_for_sandwich_nested_withClusters <- df_for_sandwich_nested %>%
# dplyr::mutate(sandwich_clusters = purrr::map(data, magrittr::extract, c("group", "school", "track")))
# The last line selected the cluster variables from the data frame of each intervention-gender combination)
DT::datatable(df_fitted)Density plot of girls and boys in different educational tracks and schools
df %>% dplyr::select(mvpaAccelerometer_T1, girl, trackSchool) %>%
group_by(girl, trackSchool) %>%
summarise(mean = mean(mvpaAccelerometer_T1, na.rm = TRUE),
median = median(mvpaAccelerometer_T1, na.rm = TRUE),
max = max(mvpaAccelerometer_T1, na.rm = TRUE),
min = min(mvpaAccelerometer_T1, na.rm = TRUE),
sd = sd(mvpaAccelerometer_T1, na.rm = TRUE),
n = n()) %>%
DT::datatable() #papaja::apa_table()
df %>% dplyr::select(mvpaAccelerometer_T1, girl, trackSchool) %>%
group_by(girl) %>%
summarise(mean = mean(mvpaAccelerometer_T1, na.rm = TRUE),
median = median(mvpaAccelerometer_T1, na.rm = TRUE),
max = max(mvpaAccelerometer_T1, na.rm = TRUE),
min = min(mvpaAccelerometer_T1, na.rm = TRUE),
sd = sd(mvpaAccelerometer_T1, na.rm = TRUE),
n = n()) %>%
DT::datatable() #papaja::apa_table()
plot1 <- df %>% dplyr::select(id,
trackSchool = trackSchool,
girl,
PA = mvpaAccelerometer_T1) %>%
dplyr::filter(!is.na(trackSchool), !grepl('Other|NANA|BA4|HRC2|Nur2', trackSchool)) %>% # Drop categories with just few participants
ggplot2::ggplot(aes(y = trackSchool)) +
ggridges::geom_density_ridges2(aes(x = PA, colour = "black",
fill = paste(trackSchool, girl)),
scale = 1,
alpha = 0.6, size = 0.25,
# from = 0, to = 450,
jittered_points=TRUE, point_shape=21,
point_fill="black") +
labs(x = "",
y = "") +
scale_y_discrete(expand = c(0.01, 0)) +
scale_x_continuous(expand = c(0.01, 0)) +
ggridges::scale_fill_cyclical(
labels = c('BA1 boy' = "Boy", 'BA1 girl' = "Girl"),
values = viridis::viridis(4, end = 0.8)[c(1, 3)],
name = "", guide = guide_legend(override.aes = list(alpha = 1))) +
ggridges::scale_colour_cyclical(values = "black") +
ggridges::theme_ridges(grid = FALSE) +
theme(legend.position="bottom", axis.text=element_text(size=10))
plot1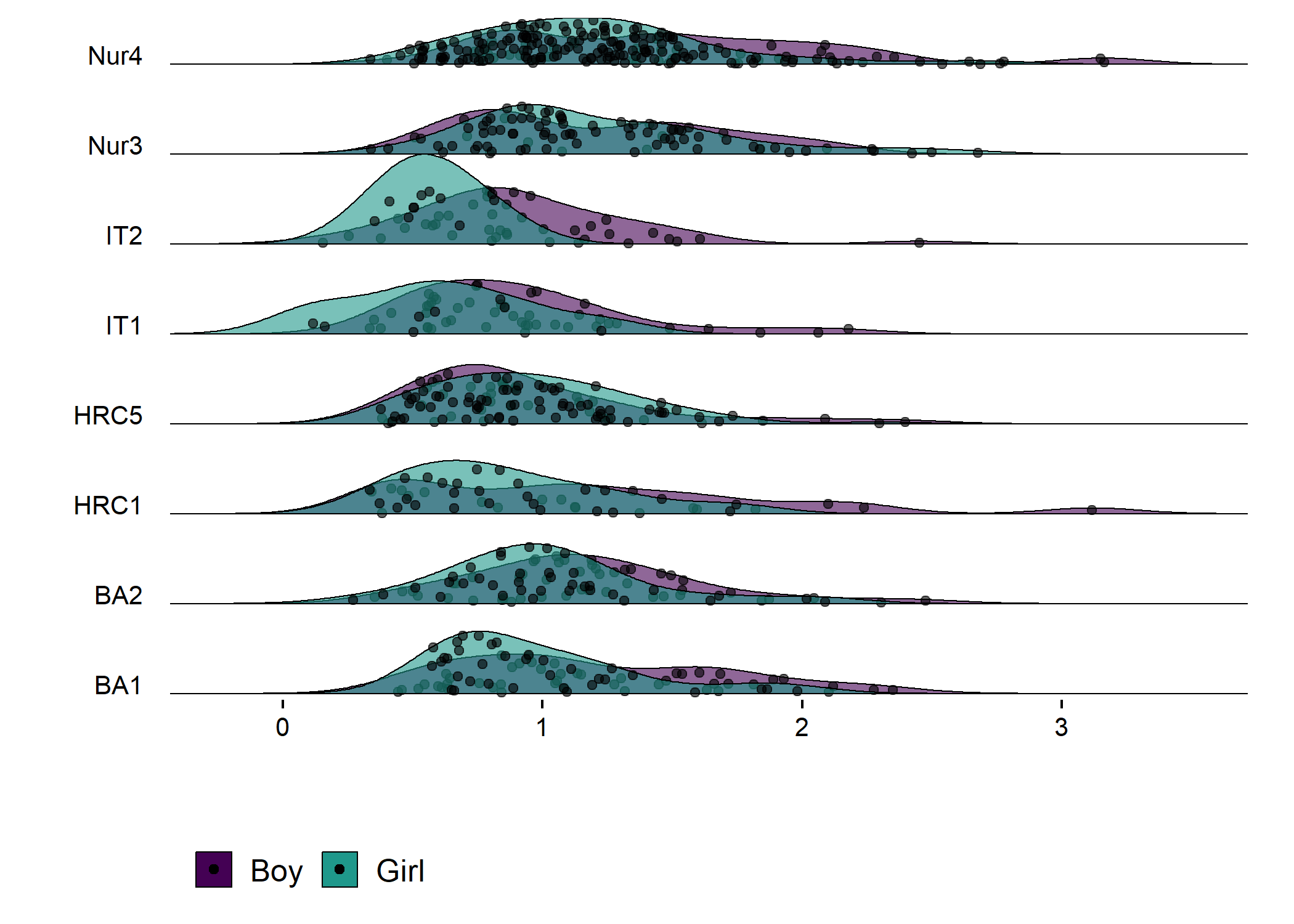
Density plot by track
plot1 <- df %>% dplyr::select(id,
track = track,
girl,
PA = mvpaAccelerometer_T1) %>%
dplyr::filter(!is.na(track), track != "Other") %>% # Drop category "Other"
dplyr::mutate(track = factor(track, levels = c("IT", "BA", "HRC", "Nur")),
track = recode_factor(track, "IT" = "IT", "BA" = "BA",
"HRC" = "HRC", "Nur" = "Nur")) %>%
ggplot2::ggplot(aes(y = track)) +
ggridges::geom_density_ridges2(aes(x = PA, colour = "black",
fill = paste(track, girl),
point_color = girl,
point_fill = girl,
point_shape = girl),
scale = .75, alpha = 0.6, size = 0.25,
position = position_raincloud(width = 0.05, height = 0.15),
# from = 0, to = 450,
jittered_points = TRUE,
point_size = 1) +
labs(x = NULL,
y = NULL) +
scale_y_discrete(expand = c(0.1, 0)) +
scale_x_continuous(expand = c(0.01, 0)) +
ggridges::scale_fill_cyclical(
labels = c('BA boy' = "Boy", 'BA girl' = "Girl"),
values = viridis::viridis(4, end = 0.8)[c(1, 3)],
name = "",
guide = guide_legend(override.aes = list(alpha = 1,
point_shape = c(3, 4),
point_size = 2))) +
ggridges::scale_colour_cyclical(values = "black") +
ggridges::theme_ridges(grid = FALSE) +
# ggplot2::scale_fill_manual(values = c("#A0FFA0", "#A0A0FF")) +
ggridges::scale_discrete_manual(aesthetics = "point_color",
values = viridis::viridis(4, end = 0.8)[c(3, 1)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_fill",
values = viridis::viridis(4, end = 0.8)[c(3, 1)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_shape",
values = c(4, 3),
guide = "none") +
papaja::theme_apa() +
theme(legend.position = "bottom")
PA_trackgirl_diamonds_girl_hours <- PA_trackgirl_diamonds_girl %>%
dplyr::mutate_if(is.numeric, ~./60)
PA_trackgirl_diamonds_boy_hours <- PA_trackgirl_diamonds_boy %>%
dplyr::mutate_if(is.numeric, ~./60)
plot1 <- plot1 +
userfriendlyscience::diamondPlot(PA_trackgirl_diamonds_girl_hours,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[3],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15)) + # specify y-position; option to move the diamond
userfriendlyscience::diamondPlot(PA_trackgirl_diamonds_boy_hours,
returnLayerOnly = TRUE,
lineColor = "black",
linetype = "solid",
color=viridis::viridis(4, end = 0.8)[1],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15))
# Draw plot with intervention and control densities
plot2 <- df %>% dplyr::select(id,
track = track,
intervention,
PA = mvpaAccelerometer_T1) %>%
dplyr::filter(!is.na(track), track != "Other") %>% # Drop category "Other"
dplyr::mutate(track = factor(track, levels = c("IT", "BA", "HRC", "Nur")),
track = recode_factor(track, "IT" = "IT", "BA" = "BA",
"HRC" = "HRC", "Nur" = "Nur")) %>%
ggplot2::ggplot(aes(y = track)) +
ggridges::geom_density_ridges2(aes(x = PA, colour = "black",
fill = paste(track, intervention),
point_color = intervention,
point_fill = intervention,
point_shape = intervention),
scale = .75, alpha = 0.6, size = 0.25,
position = position_raincloud(width = 0.05, height = 0.15),
# from = 0, to = 450,
jittered_points = TRUE,
point_size = 1) +
labs(x = NULL,
y = NULL) +
scale_y_discrete(expand = c(0.1, 0), labels = NULL) +
scale_x_continuous(expand = c(0.01, 0)) +
ggridges::scale_fill_cyclical(
labels = c('BA 0' = "Control", 'BA 1' = "Intervention"),
values = viridis::viridis(4, end = 0.8)[c(2, 4)],
name = "",
guide = guide_legend(override.aes = list(alpha = 1,
point_shape = c(4, 3),
point_size = 2))) +
ggridges::scale_colour_cyclical(values = "black") +
ggridges::theme_ridges(grid = FALSE) +
# ggplot2::scale_fill_manual(values = c("#A0FFA0", "#A0A0FF")) +
ggridges::scale_discrete_manual(aesthetics = "point_color",
values = viridis::viridis(4, end = 0.8)[c(2, 4)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_fill",
values = viridis::viridis(4, end = 0.8)[c(2, 4)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_shape",
values = c(4, 3),
guide = "none") +
papaja::theme_apa() +
theme(legend.position = "bottom")
PA_trackintervention_diamonds_intervention_hours <- PA_trackintervention_diamonds_intervention %>%
dplyr::mutate_if(is.numeric, ~./60)
PA_trackintervention_diamonds_control_hours <- PA_trackintervention_diamonds_control %>%
dplyr::mutate_if(is.numeric, ~./60)
plot2 <- plot2 +
userfriendlyscience::diamondPlot(PA_trackintervention_diamonds_intervention_hours,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[4],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15)) +
userfriendlyscience::diamondPlot(PA_trackintervention_diamonds_control_hours,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[2],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15))
plot1 + plot2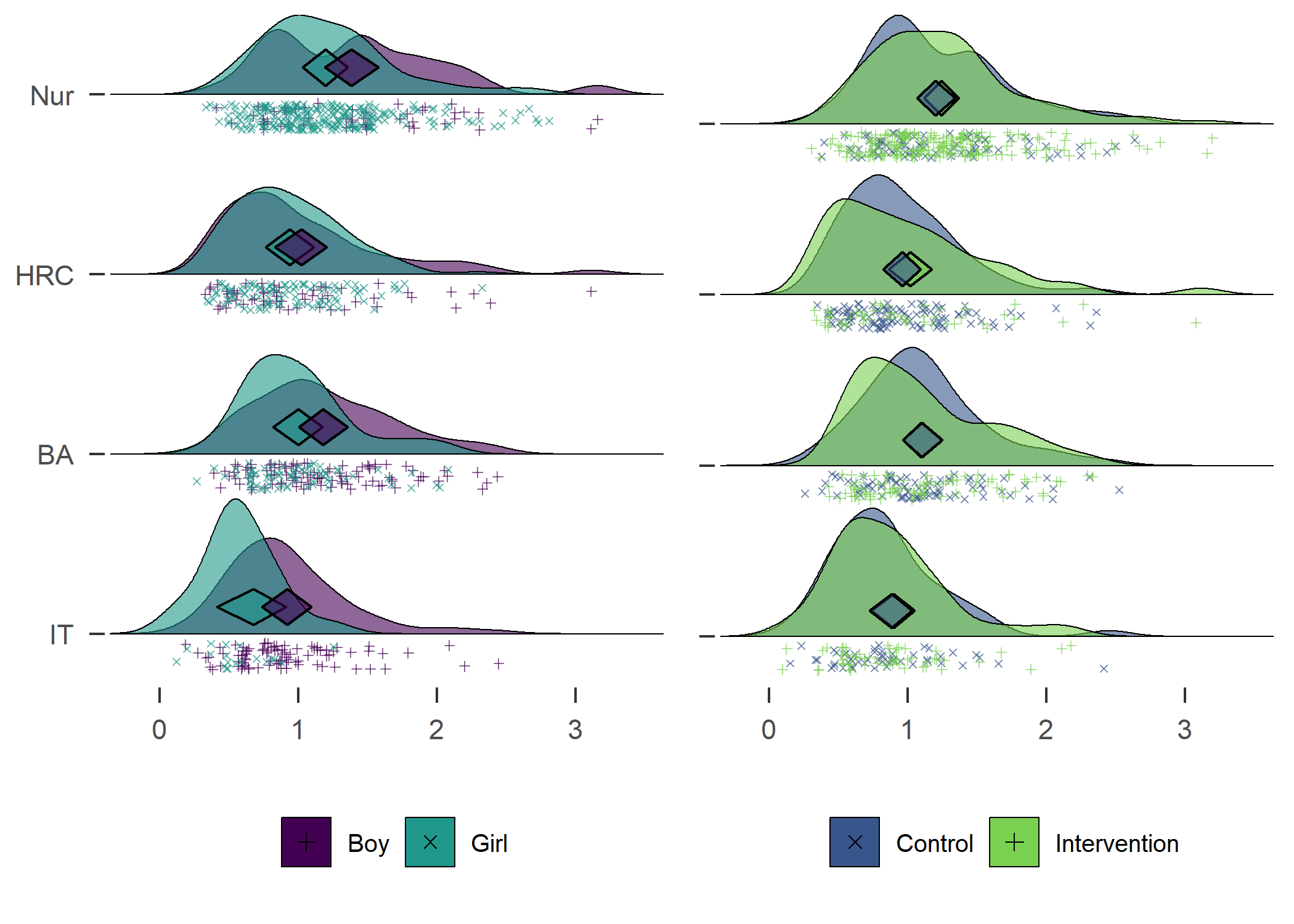
Accelerometer-measured MVPA. Nur = Practical nurse, HRC = Hotel, restaurant and catering, BA = Business and administration, IT = Information and communications technology.
Self-reported MVPA days, previous week
Description
This section reports the question on how many days the participants did MVPA during the previous week. Question was formulated as “During the last 7 days, on how many days were you physically active so, that the activity was at least moderately vigorous, and at least 30 minutes of activity was accumulated during the day? Choose the correct option”.The scale allowed discrete values from zero to seven.
Data preparation
Description
The tab “Information on data preparation” of this section present information on data preparation for the plot
Information on data preparation
Code chunk below prepares data.
# m_padaysLastweek_trackgirl <- brms::bf(padaysLastweek_T1 ~ (1 | track:girl)) %>%
# brms::brm(., data = df, chains = 4, iter = 4000, control = list(adapt_delta = 0.95))
#
# brms::prior_summary(m_padaysLastweek_trackgirl, data = df)
#
# m_padaysLastweek_trackgirl
#
# b_intercept <- brms::fixef(m_padaysLastweek_trackgirl)[1]
#
# # This gives estimates only:
# padaysLastweek_trackgirl_estimates <- brms::ranef(m_padaysLastweek_trackgirl)[[1]][1:10]
#
# # The 2.5%ile:
# padaysLastweek_trackgirl_CIL <- brms::ranef(m_padaysLastweek_trackgirl)[[1]][21:30]
#
# # The 97.5%ile:
# padaysLastweek_trackgirl_CIH <- brms::ranef(m_padaysLastweek_trackgirl)[[1]][31:40]
#
# padaysLastweek_trackgirl_ci <- data.frame(padaysLastweek_trackgirl_CIL, padaysLastweek_trackgirl_estimates, padaysLastweek_trackgirl_CIH, Variable = labels(brms::ranef(m_padaysLastweek_trackgirl))) %>%
# dplyr::mutate(b_intercept = rep(b_intercept, 10)) %>%
# dplyr::mutate(padaysLastweek_trackgirl_CIL = padaysLastweek_trackgirl_CIL + b_intercept,
# padaysLastweek_trackgirl_estimates = padaysLastweek_trackgirl_estimates + b_intercept,
# padaysLastweek_trackgirl_CIH = padaysLastweek_trackgirl_CIH + b_intercept,
# track = c('BA_boy',
# 'BA_girl',
# 'HRC_boy',
# 'HRC_girl',
# 'IT_boy',
# 'IT_girl',
# 'Nur_boy',
# 'Nur_girl',
# 'Other_boy',
# 'Other_girl'
# ))
#
# padaysLastweek_trackgirl_diamonds_girl <- rbind(
# padaysLastweek_trackgirl_ci %>% dplyr::filter(track == "Nur_girl"),
# padaysLastweek_trackgirl_ci %>% dplyr::filter(track == "HRC_girl"),
# padaysLastweek_trackgirl_ci %>% dplyr::filter(track == "BA_girl"),
# padaysLastweek_trackgirl_ci %>% dplyr::filter(track == "IT_girl")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
# padaysLastweek_trackgirl_diamonds_boy <- rbind(
# padaysLastweek_trackgirl_ci %>% dplyr::filter(track == "Nur_boy"),
# padaysLastweek_trackgirl_ci %>% dplyr::filter(track == "HRC_boy"),
# padaysLastweek_trackgirl_ci %>% dplyr::filter(track == "BA_boy"),
# padaysLastweek_trackgirl_ci %>% dplyr::filter(track == "IT_boy")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
#
# m_padaysLastweek_trackintervention <- brms::bf(padaysLastweek_T1 ~ (1 | track:intervention)) %>%
# brms::brm(., data = df, chains = 4, iter = 4000, control = list(adapt_delta = 0.95))
#
# brms::prior_summary(m_padaysLastweek_trackintervention, data = df)
#
# m_padaysLastweek_trackintervention
#
# b_intercept <- brms::fixef(m_padaysLastweek_trackintervention)[1]
#
# # This gives estimates only:
# padaysLastweek_trackintervention_estimates <- brms::ranef(m_padaysLastweek_trackintervention)[[1]][1:10]
#
# # The 2.5%ile:
# padaysLastweek_trackintervention_CIL <- brms::ranef(m_padaysLastweek_trackintervention)[[1]][21:30]
#
# # The 97.5%ile:
# padaysLastweek_trackintervention_CIH <- brms::ranef(m_padaysLastweek_trackintervention)[[1]][31:40]
#
# padaysLastweek_trackintervention_ci <- data.frame(padaysLastweek_trackintervention_CIL, padaysLastweek_trackintervention_estimates, padaysLastweek_trackintervention_CIH, Variable = labels(brms::ranef(m_padaysLastweek_trackintervention))) %>%
# dplyr::mutate(b_intercept = rep(b_intercept, 10)) %>%
# dplyr::mutate(padaysLastweek_trackintervention_CIL = padaysLastweek_trackintervention_CIL + b_intercept,
# padaysLastweek_trackintervention_estimates = padaysLastweek_trackintervention_estimates + b_intercept,
# padaysLastweek_trackintervention_CIH = padaysLastweek_trackintervention_CIH + b_intercept,
# track = c('BA_control',
# 'BA_intervention',
# 'HRC_control',
# 'HRC_intervention',
# 'IT_control',
# 'IT_intervention',
# 'Nur_control',
# 'Nur_intervention',
# 'Other_control',
# 'Other_intervention'
# ))
#
# padaysLastweek_trackintervention_diamonds_intervention <- rbind(
# padaysLastweek_trackintervention_ci %>% dplyr::filter(track == "Nur_intervention"),
# padaysLastweek_trackintervention_ci %>% dplyr::filter(track == "HRC_intervention"),
# padaysLastweek_trackintervention_ci %>% dplyr::filter(track == "BA_intervention"),
# padaysLastweek_trackintervention_ci %>% dplyr::filter(track == "IT_intervention")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
# padaysLastweek_trackintervention_diamonds_control <- rbind(
# padaysLastweek_trackintervention_ci %>% dplyr::filter(track == "Nur_control"),
# padaysLastweek_trackintervention_ci %>% dplyr::filter(track == "HRC_control"),
# padaysLastweek_trackintervention_ci %>% dplyr::filter(track == "BA_control"),
# padaysLastweek_trackintervention_ci %>% dplyr::filter(track == "IT_control")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
# save(padaysLastweek_trackgirl_diamonds_girl, file = "./Rdata_files/padaysLastweek_trackgirl_diamonds_girl.Rdata")
# save(padaysLastweek_trackgirl_diamonds_boy, file = "./Rdata_files/padaysLastweek_trackgirl_diamonds_boy.Rdata")
# save(padaysLastweek_trackintervention_diamonds_intervention, file = "./Rdata_files/padaysLastweek_trackintervention_diamonds_intervention.Rdata")
# save(padaysLastweek_trackintervention_diamonds_control, file = "./Rdata_files/padaysLastweek_trackintervention_diamonds_control.Rdata")
load("./Rdata_files/padaysLastweek_trackgirl_diamonds_girl.Rdata")
load("./Rdata_files/padaysLastweek_trackgirl_diamonds_boy.Rdata")
load("./Rdata_files/padaysLastweek_trackintervention_diamonds_intervention.Rdata")
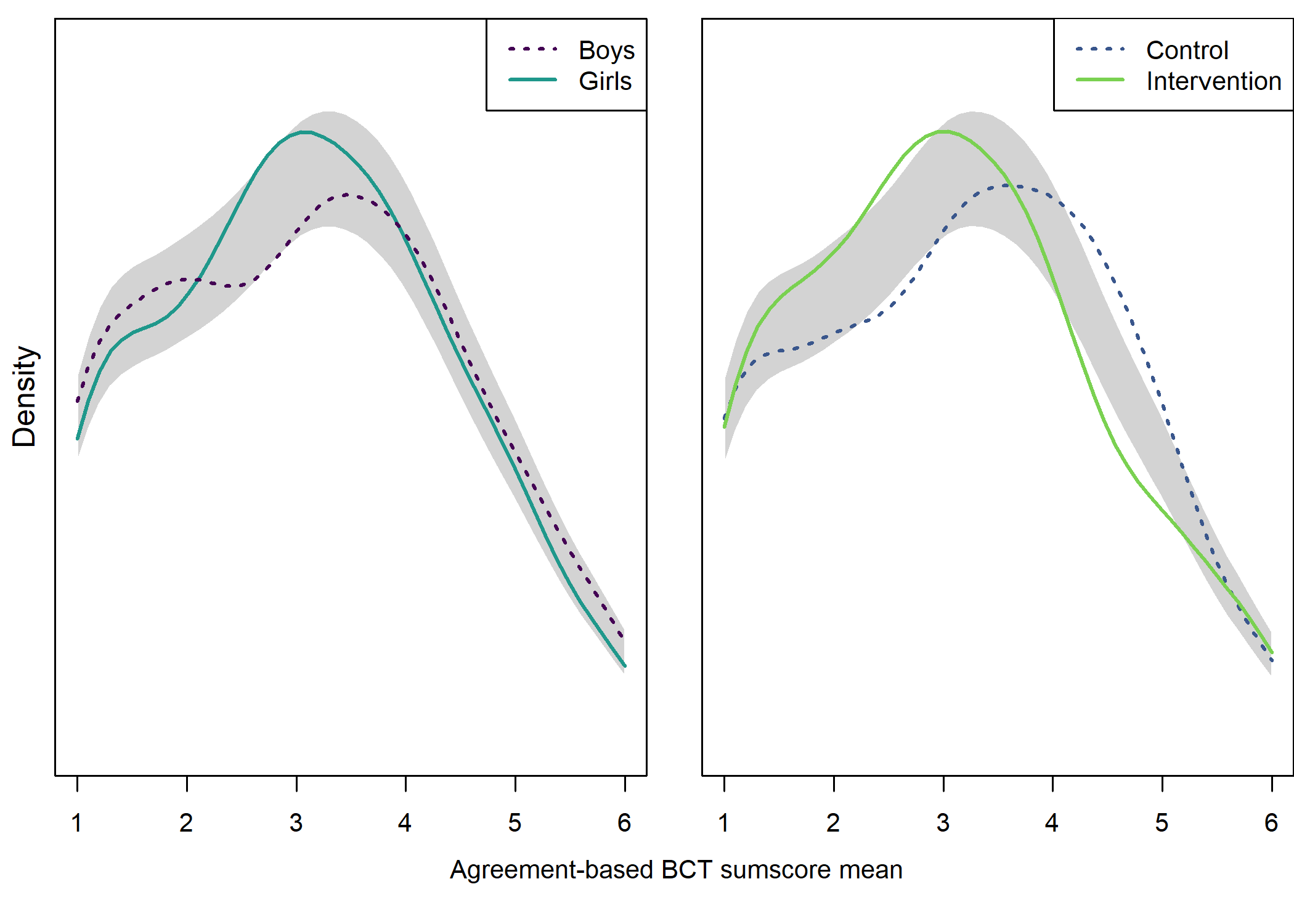
load("./Rdata_files/padaysLastweek_trackintervention_diamonds_control.Rdata")Density plot by intervention and gender
# Create data frame
densplot <- d
levels(densplot$intervention) <- c("Control", "Intervention")
levels(densplot$girl) <- recode(densplot$girl, "boy" = "Boys", "girl" = "Girls")
# This gives side-by-side plots. There's a third plot below the two, which is fake to include x-axis text.
layout(matrix(c(1, 2, 3, 3), nrow = 2, ncol = 2, byrow = TRUE),
widths = c(0.5, 0.5), heights = c(0.45, 0.05))
# Minimise whitespace; see https://stackoverflow.com/questions/15848942/how-to-reduce-space-gap-between-multiple-graphs-in-r
par(mai = c(0.3, 0.3, 0.1, 0.0))
## Girls vs. boys
# Choose only the variables needed and drop NA
dens <- densplot %>% dplyr::select(padaysLastweek_T1, girl) %>%
dplyr::filter(complete.cases(.))
# Set random number generator for reproducibility of bootstrap test of equal densities
set.seed(10)
# Make plot
sm.padaysLastweek_T1_2 <- sm.density.compare2(as.numeric(dens$padaysLastweek_T1),
as.factor(dens$girl),
model = "equal",
xlab = "", ylab = "",
col = viridis::viridis(4, end = 0.8)[c(3, 1)],
lty = c(1, 3), yaxt = "n",
bandcol = 'LightGray',
lwd = c(2, 2),
xlim = c(0, 7))
##
## Test of equal densities: p-value = 0
legend("topright", levels(dens$girl), col = viridis::viridis(4, end = 0.8)[c(1, 3)], lty = c(3,1), lwd = (c(2,2)))
mtext(side = 2, "Density", line = 0.5)
## Intervention vs. control
# Choose only the variables needed and drop NA
dens <- densplot %>% dplyr::select(padaysLastweek_T1, intervention) %>%
dplyr::filter(complete.cases(.))
# Set random number generator for reproducibility of bootstrap test of equal densities
set.seed(10)
# Make plot
sm.padaysLastweek_T1_2 <- sm.density.compare2(as.numeric(dens$padaysLastweek_T1),
as.factor(dens$intervention),
model = "equal",
xlab = "", ylab = "",
col = viridis::viridis(4, end = 0.8)[c(2, 4)],
lty = c(3, 1), yaxt = "n",
bandcol = 'LightGray',
lwd = c(2, 2),
xlim = c(0, 7))
##
## Test of equal densities: p-value = 0.09
legend("topright", levels(dens$intervention), col = viridis::viridis(4, end = 0.8)[c(2, 4)], lty=c(3,1), lwd=(c(2,2)))
# Create x-axis label. See https://stackoverflow.com/questions/11198767/how-to-annotate-across-or-between-plots-in-multi-plot-panels-in-r
par(mar = c(0,0,0,0))
plot(1, 1, type = "n", frame.plot = FALSE, axes = FALSE) # Fake plot for x-axis label
text(x = 1.02, y = 1.3, labels = "Self-reported number of days with >30 MVPA min previous week", pos = 1)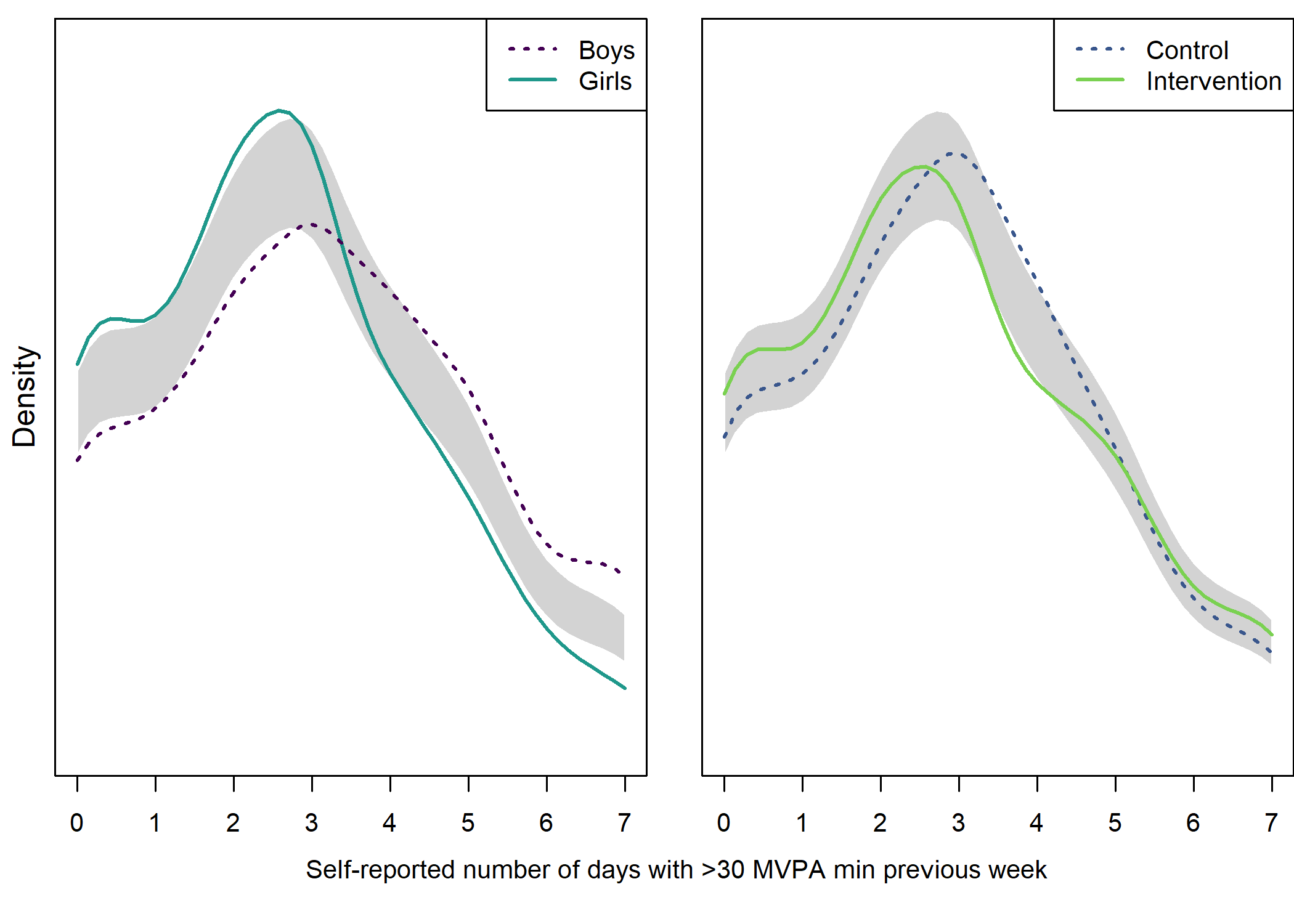
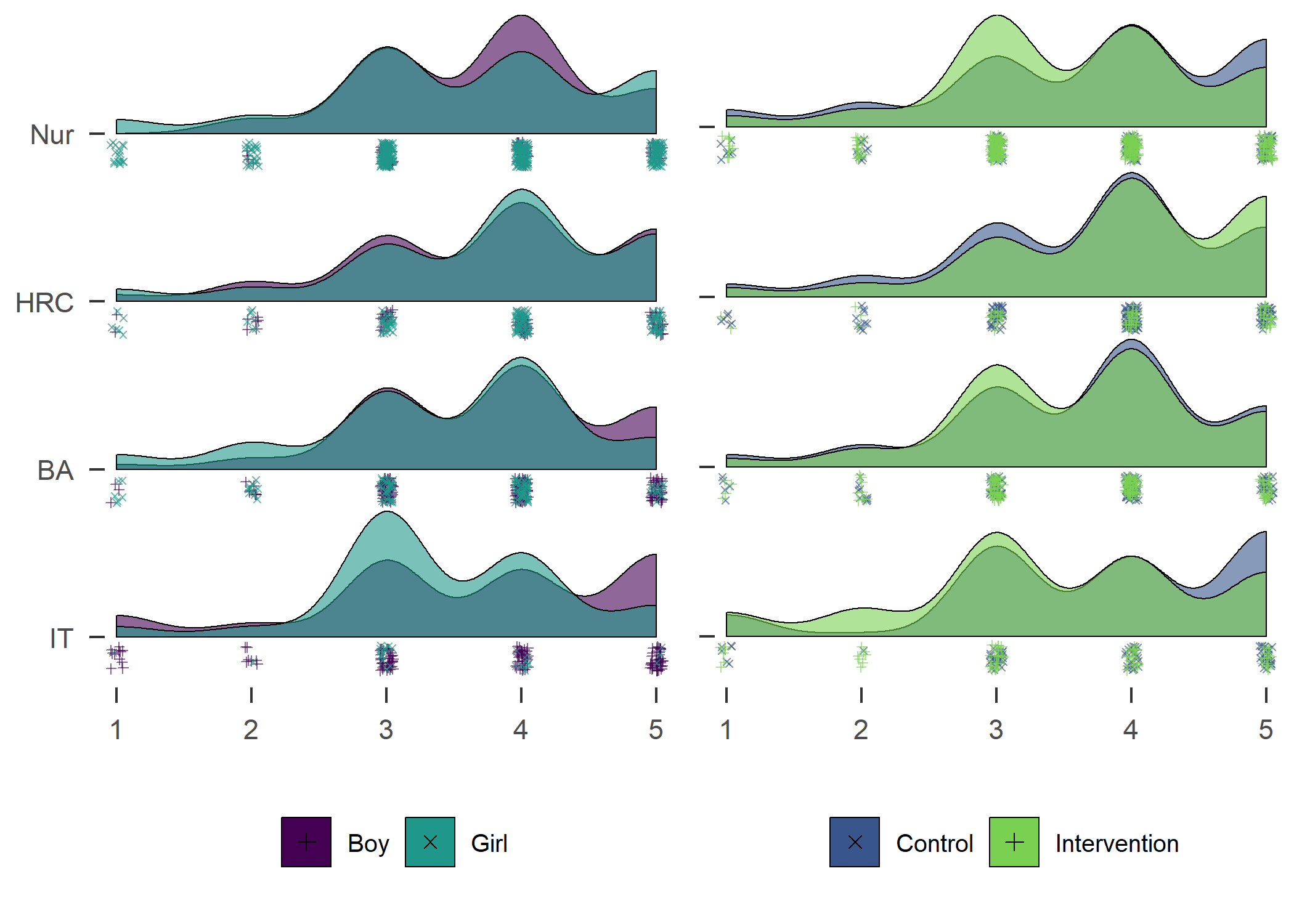
From the figure, we can see that there were more boys reporting a high number of MVPA days, and fewer boys reported low numbers. This effect was consistent among educational tracks. Boys and girls, as well as different educational tracks, differed largely in the types of PA they reported having engaged in during the previous month (see tab Tables and Estimators, at “Types of self-reported exercise”).
Density plot by track
plot1 <- df %>% dplyr::select(id,
track = track,
girl,
PA = padaysLastweek_T1) %>%
dplyr::filter(!is.na(track), track != "Other") %>% # Drop category "Other"
dplyr::mutate(track = factor(track, levels = c("IT", "BA", "HRC", "Nur")),
track = recode_factor(track, "IT" = "IT", "BA" = "BA",
"HRC" = "HRC", "Nur" = "Nur")) %>%
ggplot2::ggplot(aes(y = track)) +
ggridges::geom_density_ridges2(aes(x = PA, colour = "black",
fill = paste(track, girl),
point_color = girl,
point_fill = girl,
point_shape = girl),
scale = .6, alpha = 0.6, size = 0.25,
position = position_raincloud(width = 0.2, height = 0.4),
# from = 0, to = 450,
jittered_points = TRUE,
point_size = 1,
limits = c(0, 7)) +
labs(x = NULL,
y = NULL) +
scale_y_discrete(expand = c(0.1, 0)) +
scale_x_continuous(expand = c(0.01, 0), breaks = 0:7, limits = c(0, 7)) +
ggridges::scale_fill_cyclical(
labels = c('BA boy' = "Boy", 'BA girl' = "Girl"),
values = viridis::viridis(4, end = 0.8)[c(1, 3)],
name = "",
guide = guide_legend(override.aes = list(alpha = 1,
point_shape = c(3, 4),
point_size = 2))) +
ggridges::scale_colour_cyclical(values = "black") +
ggridges::theme_ridges(grid = FALSE) +
# ggplot2::scale_fill_manual(values = c("#A0FFA0", "#A0A0FF")) +
ggridges::scale_discrete_manual(aesthetics = "point_color",
values = viridis::viridis(4, end = 0.8)[c(3, 1)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_fill",
values = viridis::viridis(4, end = 0.8)[c(3, 1)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_shape",
values = c(4, 3),
guide = "none") +
papaja::theme_apa() +
theme(legend.position = "bottom")
plot1 <- plot1 +
userfriendlyscience::diamondPlot(padaysLastweek_trackgirl_diamonds_girl,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[3],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15)) + # specify y-position; option to move the diamond
userfriendlyscience::diamondPlot(padaysLastweek_trackgirl_diamonds_boy,
returnLayerOnly = TRUE,
lineColor = "black",
linetype = "solid",
color=viridis::viridis(4, end = 0.8)[1],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15))
# Draw plot with intervention and control densities
plot2 <- df %>% dplyr::select(id,
track = track,
intervention,
PA = padaysLastweek_T1) %>%
dplyr::filter(!is.na(track), track != "Other") %>% # Drop category "Other"
dplyr::mutate(track = factor(track, levels = c("IT", "BA", "HRC", "Nur")),
track = recode_factor(track, "IT" = "IT", "BA" = "BA",
"HRC" = "HRC", "Nur" = "Nur")) %>%
ggplot2::ggplot(aes(y = track)) +
ggridges::geom_density_ridges2(aes(x = PA, colour = "black",
fill = paste(track, intervention),
point_color = intervention,
point_fill = intervention,
point_shape = intervention),
scale = .6, alpha = 0.6, size = 0.25,
position = position_raincloud(width = 0.2, height = 0.4),
# from = 0, to = 450,
jittered_points = TRUE,
point_size = 1,
limits = c(0, 7)) +
labs(x = NULL,
y = NULL) +
scale_y_discrete(expand = c(0.1, 0), labels = NULL) +
scale_x_continuous(expand = c(0.01, 0), breaks = 0:7, limits = c(0, 7)) +
ggridges::scale_fill_cyclical(
labels = c('BA 0' = "Control", 'BA 1' = "Intervention"),
values = viridis::viridis(4, end = 0.8)[c(2, 4)],
name = "",
guide = guide_legend(override.aes = list(alpha = 1,
point_shape = c(4, 3),
point_size = 2))) +
ggridges::scale_colour_cyclical(values = "black") +
ggridges::theme_ridges(grid = FALSE) +
# ggplot2::scale_fill_manual(values = c("#A0FFA0", "#A0A0FF")) +
ggridges::scale_discrete_manual(aesthetics = "point_color",
values = viridis::viridis(4, end = 0.8)[c(2, 4)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_fill",
values = viridis::viridis(4, end = 0.8)[c(2, 4)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_shape",
values = c(4, 3),
guide = "none") +
papaja::theme_apa() +
theme(legend.position = "bottom")
plot2 <- plot2 +
userfriendlyscience::diamondPlot(padaysLastweek_trackintervention_diamonds_intervention,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[4],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15)) +
userfriendlyscience::diamondPlot(padaysLastweek_trackintervention_diamonds_control,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[2],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15))
plot1 + plot2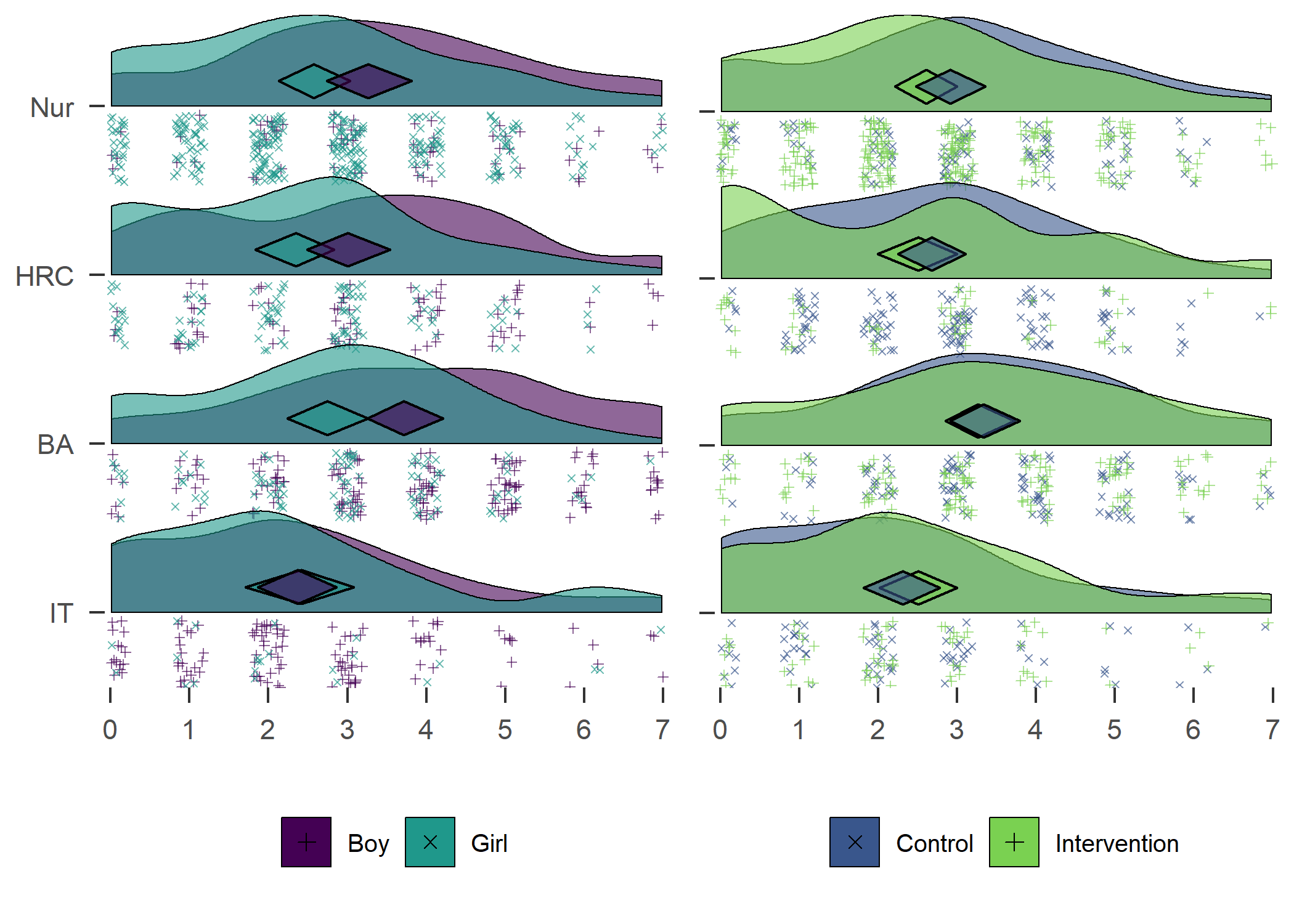
Self-reported number of days with > 30 MVPA min previous week. Nur = Practical nurse, HRC = Hotel, restaurant and catering, BA = Business and administration, IT = Information and communications technology.
Histogram 1
plotboys <- df %>% dplyr::select(id,
track = track,
girl,
padaysLastweek_T1) %>%
dplyr::filter(!is.na(track), track != "Other", girl == "boy") %>% # Drop category "Other"
dplyr::mutate(track = factor(track, levels = c("IT", "BA", "HRC", "Nur")),
track = recode_factor(track, "IT" = "IT", "BA" = "BA",
"HRC" = "HRC", "Nur" = "Nur")) %>%
ggplot2::ggplot(aes(y = track)) +
ggridges::geom_density_ridges2(aes(x = padaysLastweek_T1, fill = girl), stat = "binline", binwidth = 1, scale = 0.95, alpha = 0.6) +
scale_x_continuous(breaks = c(0:7), expand = c(0, 0), name = NULL) +
scale_y_discrete(expand = c(0.01, 0), name = "") +
ggridges::scale_fill_cyclical(values = viridis::viridis(4, end = 0.8)[1]) +
labs(title = "Boys") +
guides(y = "none") +
ggridges::theme_ridges(grid = FALSE) +
theme(axis.title.x = element_text(hjust = 0.5),
axis.title.y = element_text(hjust = 0.5),
plot.title = element_text(hjust = 0.5, size = 10),
axis.text=element_text(size=10)) +
coord_cartesian(xlim = c(-0.5, 7.5))
plotboys <- plotboys +
userfriendlyscience::diamondPlot(padaysLastweek_trackgirl_diamonds_girl,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[3],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15)) + # specify y-position; option to move the diamond
userfriendlyscience::diamondPlot(padaysLastweek_trackgirl_diamonds_boy,
returnLayerOnly = TRUE,
lineColor = "black",
linetype = "solid",
color=viridis::viridis(4, end = 0.8)[1],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15))
plotgirls <- df %>% dplyr::select(id,
track = track,
girl,
padaysLastweek_T1) %>%
dplyr::filter(!is.na(track), track != "Other", girl == "girl") %>% # Drop category "Other"
dplyr::mutate(track = factor(track, levels = c("IT", "BA", "HRC", "Nur")),
track = recode_factor(track, "IT" = "IT", "BA" = "BA",
"HRC" = "HRC", "Nur" = "Nur")) %>%
ggplot2::ggplot(aes(y = track)) +
ggridges::geom_density_ridges2(aes(x = padaysLastweek_T1, fill = girl), stat = "binline", binwidth = 1, scale = 0.95) +
scale_x_continuous(breaks = c(0:7), expand = c(0, 0), name = NULL) +
scale_y_discrete(expand = c(0.01, 0), name = "", labels = NULL) +
ggridges::scale_fill_cyclical(values = viridis::viridis(4, end = 0.8)[3]) +
labs(title = "Girls") +
guides(y = "none") +
ggridges::theme_ridges(grid = FALSE) +
theme(axis.title.x = element_text(hjust = 0.5),
axis.title.y = element_text(hjust = 0.5),
plot.title = element_text(hjust = 0.5, size = 10),
axis.text=element_text(size=10)) +
coord_cartesian(xlim = c(-0.5, 7.5))
plotgirls <- plotgirls +
userfriendlyscience::diamondPlot(padaysLastweek_trackgirl_diamonds_girl,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[3],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15)) + # specify y-position; option to move the diamond
userfriendlyscience::diamondPlot(padaysLastweek_trackgirl_diamonds_boy,
returnLayerOnly = TRUE,
lineColor = "black",
linetype = "solid",
color=viridis::viridis(4, end = 0.8)[1],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15))
plotcontrol <- df %>% dplyr::select(id,
track = track,
intervention,
padaysLastweek_T1) %>%
dplyr::filter(!is.na(track), track != "Other", intervention == "0") %>% # Drop category "Other"
dplyr::mutate(track = factor(track, levels = c("IT", "BA", "HRC", "Nur")),
track = recode_factor(track, "IT" = "IT", "BA" = "BA",
"HRC" = "HRC", "Nur" = "Nur")) %>%
ggplot2::ggplot(aes(y = track)) +
ggridges::geom_density_ridges2(aes(x = padaysLastweek_T1, fill = intervention), stat = "binline", binwidth = 1, scale = 0.95) +
scale_x_continuous(breaks = c(0:7), expand = c(0, 0), name = NULL) +
scale_y_discrete(expand = c(0.01, 0), name = "", labels = NULL) +
ggridges::scale_fill_cyclical(values = viridis::viridis(4, end = 0.8)[2]) +
labs(title = "Control") +
guides(y = "none") +
ggridges::theme_ridges(grid = FALSE) +
theme(axis.title.x = element_text(hjust = 0.5),
axis.title.y = element_text(hjust = 0.5),
plot.title = element_text(hjust = 0.5, size = 10),
axis.text=element_text(size=10)) +
coord_cartesian(xlim = c(-0.5, 7.5))
plotcontrol <- plotcontrol +
userfriendlyscience::diamondPlot(padaysLastweek_trackintervention_diamonds_intervention,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[4],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15)) +
userfriendlyscience::diamondPlot(padaysLastweek_trackintervention_diamonds_control,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[2],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15))
plotintervention <- df %>% dplyr::select(id,
track = track,
intervention,
padaysLastweek_T1) %>%
dplyr::filter(!is.na(track), track != "Other", intervention == "1") %>% # Drop category "Other"
dplyr::mutate(track = factor(track, levels = c("IT", "BA", "HRC", "Nur")),
track = recode_factor(track, "IT" = "IT", "BA" = "BA",
"HRC" = "HRC", "Nur" = "Nur")) %>%
ggplot2::ggplot(aes(y = track)) +
ggridges::geom_density_ridges2(aes(x = padaysLastweek_T1, fill = intervention), stat = "binline", binwidth = 1, scale = 0.95) +
scale_x_continuous(breaks = c(0:7), expand = c(0, 0), name = NULL) +
scale_y_discrete(expand = c(0.01, 0), name = "", labels = NULL) +
ggridges::scale_fill_cyclical(values = viridis::viridis(4, end = 0.8)[4]) +
labs(title = "Intervention") +
guides(y = "none") +
ggridges::theme_ridges(grid = FALSE) +
theme(axis.title.x = element_text(hjust = 0.5),
axis.title.y = element_text(hjust = 0.5),
plot.title = element_text(hjust = 0.5, size = 10),
axis.text=element_text(size=10)) +
coord_cartesian(xlim = c(-0.5, 7.5))
plotintervention <- plotintervention +
userfriendlyscience::diamondPlot(padaysLastweek_trackintervention_diamonds_intervention,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[4],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15)) +
userfriendlyscience::diamondPlot(padaysLastweek_trackintervention_diamonds_control,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[2],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15))
plotboys + plotgirls + plotcontrol + plotintervention + plot_layout(ncol = 4)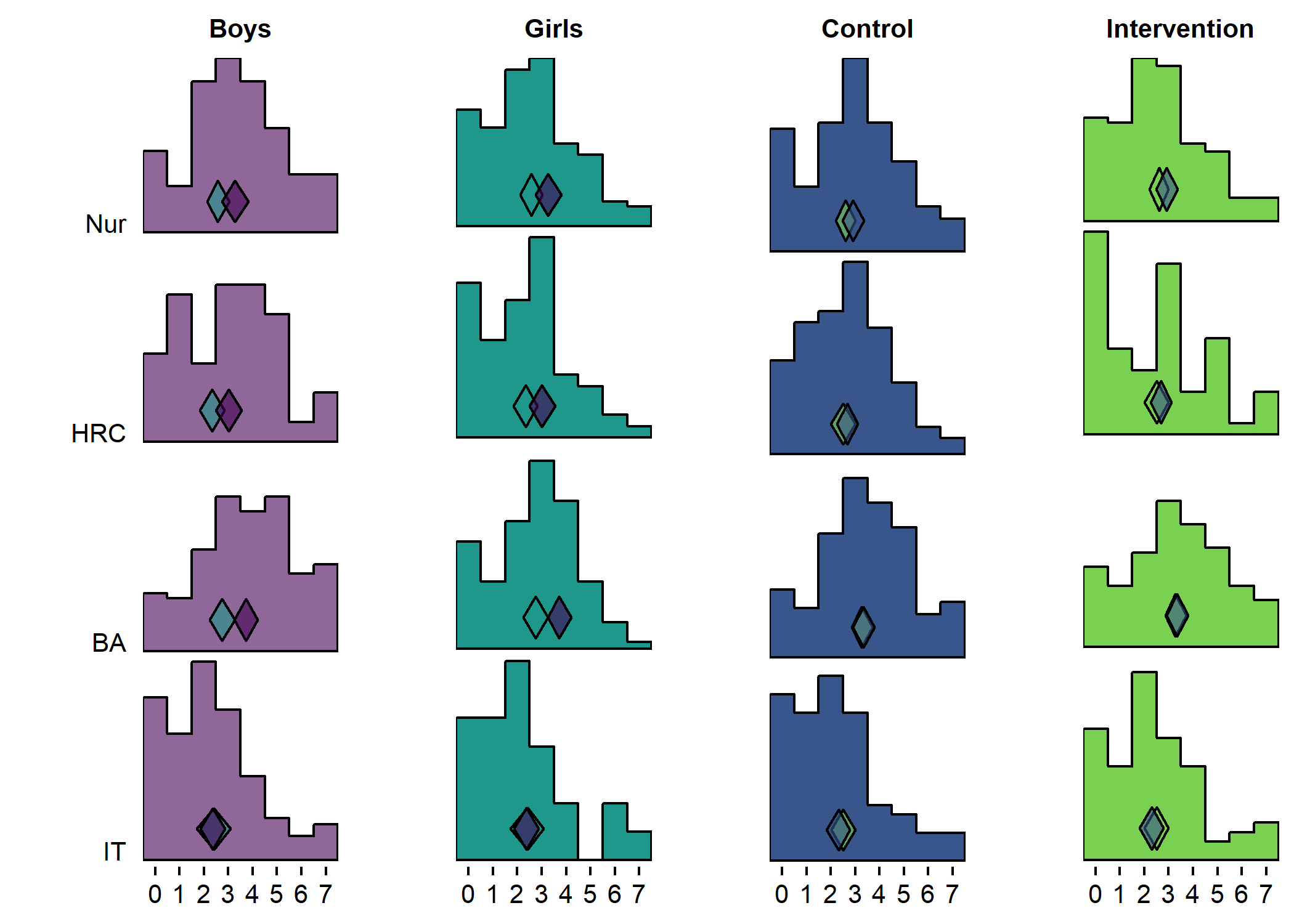
Histogram 2
hist_padaysLastweek_girl_count <- df %>% dplyr::select(id,
track = track,
girl,
padaysLastweek_T1) %>%
dplyr::filter(!is.na(track), track != "Other") %>% # Drop category "Other"
dplyr::mutate(track = factor(track, levels = c("IT", "BA", "HRC", "Nur")),
track = recode_factor(track, "IT" = "IT", "BA" = "BA",
"HRC" = "HRC", "Nur" = "Nur"),
padaysLastweek_T1 = as.integer(padaysLastweek_T1)) %>%
ggplot(aes(x=padaysLastweek_T1, fill=girl)) +
geom_histogram(binwidth=1, position="dodge", color = "black") +
scale_x_continuous(breaks = 0:7) +
labs(x = NULL) +
scale_fill_manual(values = viridis::viridis(4, end = 0.8)[c(3, 1)],
name = NULL) +
facet_wrap("track") +
theme(legend.position = "bottom")
hist_padaysLastweek_girl_density <- df %>% dplyr::select(id,
track = track,
girl,
padaysLastweek_T1) %>%
dplyr::filter(!is.na(track), track != "Other") %>% # Drop category "Other"
dplyr::mutate(track = factor(track, levels = c("IT", "BA", "HRC", "Nur")),
track = recode_factor(track, "IT" = "IT", "BA" = "BA",
"HRC" = "HRC", "Nur" = "Nur"),
padaysLastweek_T1 = as.integer(padaysLastweek_T1)) %>%
ggplot(aes(x=padaysLastweek_T1, y = ..density.., fill=girl)) +
geom_histogram(binwidth=1, position="dodge", color = "black") +
scale_x_continuous(breaks = 0:7) +
labs(x = NULL) +
scale_fill_manual(values = viridis::viridis(4, end = 0.8)[c(3, 1)], name = NULL,
guide = FALSE) +
facet_wrap("track")
hist_padaysLastweek_girl_count + hist_padaysLastweek_girl_density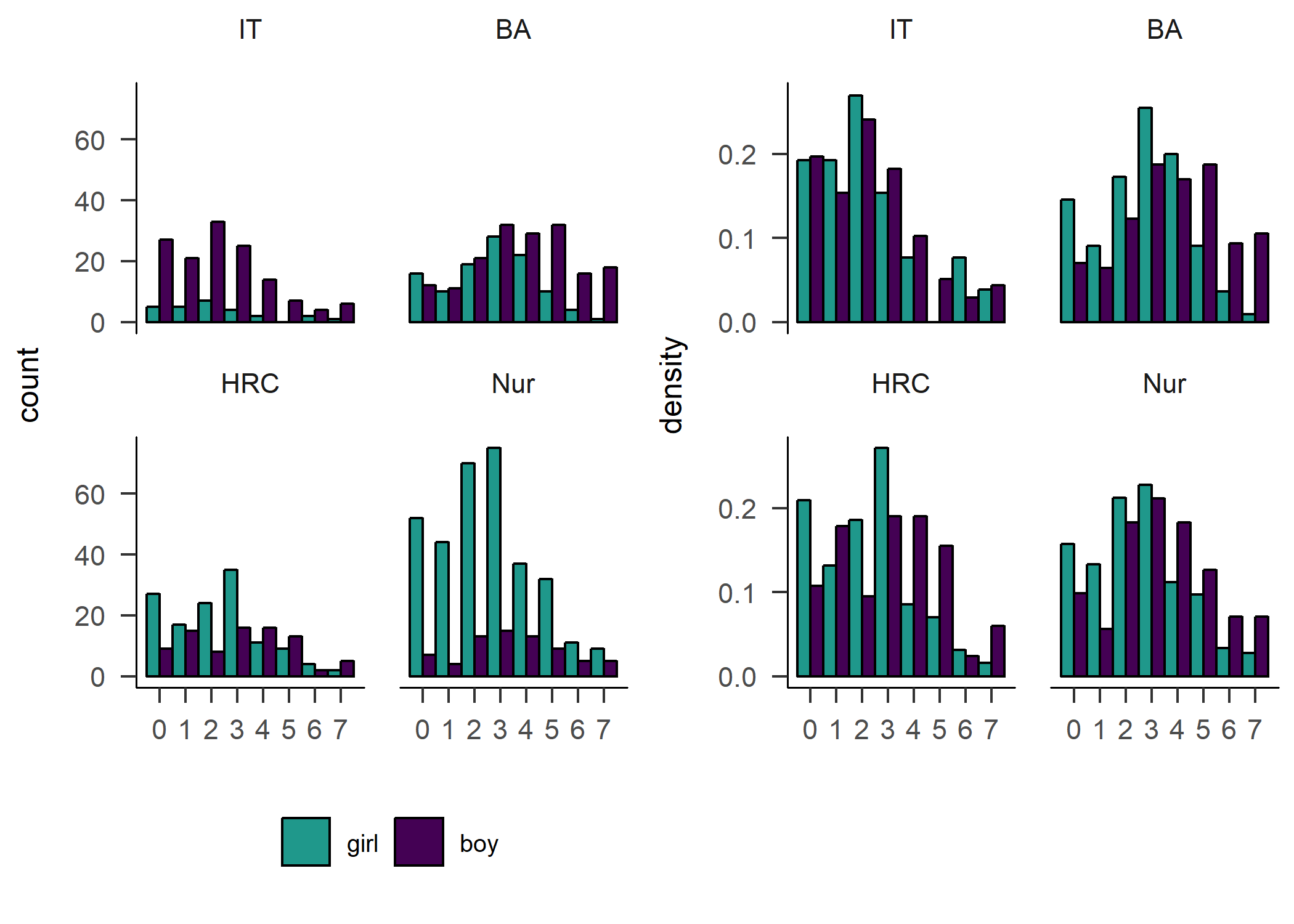
hist_padaysLastweek_intervention_count <- df %>% dplyr::select(id,
track = track,
intervention,
padaysLastweek_T1) %>%
dplyr::filter(!is.na(track), track != "Other") %>% # Drop category "Other"
dplyr::mutate(track = factor(track, levels = c("IT", "BA", "HRC", "Nur")),
track = recode_factor(track, "IT" = "IT", "BA" = "BA",
"HRC" = "HRC", "Nur" = "Nur"),
padaysLastweek_T1 = as.integer(padaysLastweek_T1)) %>%
ggplot(aes(x=padaysLastweek_T1, fill=intervention)) +
geom_histogram(binwidth = 1, position = "dodge", color = "black") +
scale_x_continuous(breaks = 0:7) +
labs(x = NULL) +
scale_fill_manual(values = viridis::viridis(4, end = 0.8)[c(2, 4)],
name = NULL,
labels = c("0" = "control", "1" = "intervention")) +
facet_wrap("track") +
theme(legend.position = "bottom")
hist_padaysLastweek_intervention_density <- df %>% dplyr::select(id,
track = track,
intervention,
padaysLastweek_T1) %>%
dplyr::filter(!is.na(track), track != "Other") %>% # Drop category "Other"
dplyr::mutate(track = factor(track, levels = c("IT", "BA", "HRC", "Nur")),
track = recode_factor(track, "IT" = "IT", "BA" = "BA",
"HRC" = "HRC", "Nur" = "Nur"),
padaysLastweek_T1 = as.integer(padaysLastweek_T1)) %>%
ggplot(aes(x=padaysLastweek_T1, y = ..density.., fill=intervention)) +
geom_histogram(binwidth = 1, position = "dodge", color = "black") +
scale_x_continuous(breaks = 0:7) +
labs(x = NULL) +
scale_fill_manual(values = viridis::viridis(4, end = 0.8)[c(2, 4)],
guide = FALSE) +
facet_wrap("track")
hist_padaysLastweek_intervention_count + hist_padaysLastweek_intervention_density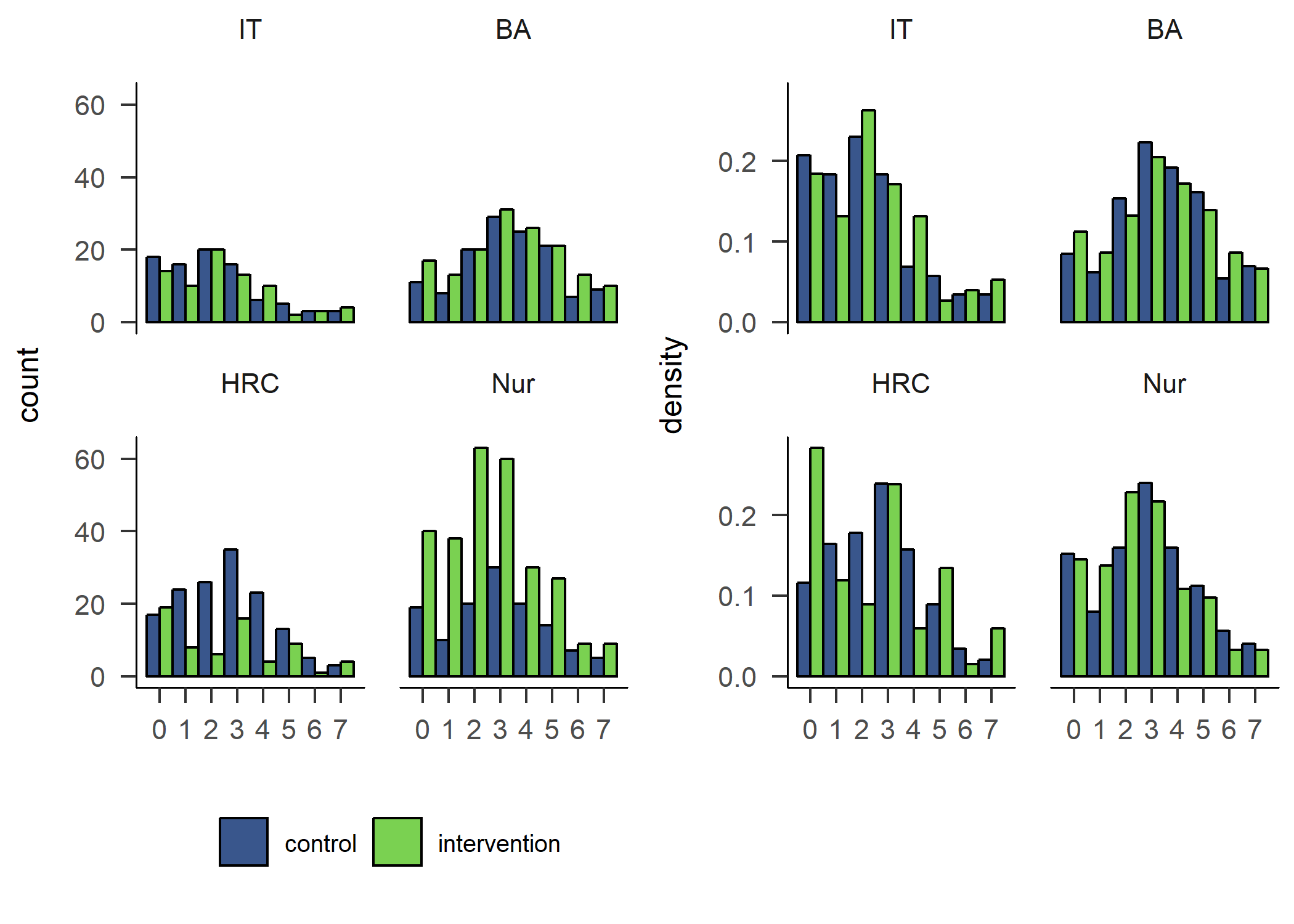
The number of days participants reported doing PA on, suggested a different picture of difference between genders in activity. Boys reported being more active in all but the IT track. The effect is also shown in the following variable on self-reported MVPA time.
Self-reported MVPA time
This question was “During the last 7 days, how many hours of the previously described [i.e. moderate-to-vigorous] physical activity was accumulated during your free time? Give your answer to the nearest 30 minutes.” The answer was given by inputing hours and minutes, the latter field allowing for either zero or 30 minutes to be inputed.
Density plot by intervention and gender
# Create data frame
densplot <- d
levels(densplot$intervention) <- c("Control", "Intervention")
levels(densplot$girl) <- recode(densplot$girl, "boy" = "Boys", "girl" = "Girls")
# This gives side-by-side plots. There's a third plot below the two, which is fake to include x-axis text.
layout(matrix(c(1, 2, 3, 3), nrow = 2, ncol = 2, byrow = TRUE),
widths = c(0.5, 0.5), heights = c(0.45, 0.05))
# Minimise whitespace; see https://stackoverflow.com/questions/15848942/how-to-reduce-space-gap-between-multiple-graphs-in-r
par(mai = c(0.3, 0.3, 0.1, 0.0))
## Girls vs. boys
# Choose only the variables needed and drop NA
dens <- densplot %>% dplyr::select(leisuretimeMvpaHoursLastweek_T1, girl) %>%
dplyr::filter(complete.cases(.))
# Set random number generator for reproducibility of bootstrap test of equal densities
set.seed(10)
# Make plot
sm.leisuretimeMvpaHoursLastweek_T1_2 <- sm.density.compare2(as.numeric(dens$leisuretimeMvpaHoursLastweek_T1),
as.factor(dens$girl),
model = "equal",
xlab = "", ylab = "",
col = viridis::viridis(4, end = 0.8)[c(3, 1)],
lty = c(1,3), yaxt = "n",
bandcol = 'LightGray',
lwd = (c(2,2)))
##
## Test of equal densities: p-value = 0
legend("topright", levels(dens$girl), col = viridis::viridis(4, end = 0.8)[c(1, 3)], lty = c(3,1), lwd = (c(2,2)))
mtext(side = 2, "Density", line = 0.5)
## Intervention vs. control
# Choose only the variables needed and drop NA
dens <- densplot %>% dplyr::select(leisuretimeMvpaHoursLastweek_T1, intervention) %>%
dplyr::filter(complete.cases(.))
# Set random number generator for reproducibility of bootstrap test of equal densities
set.seed(10)
# Make plot
sm.leisuretimeMvpaHoursLastweek_T1_2 <- sm.density.compare2(as.numeric(dens$leisuretimeMvpaHoursLastweek_T1),
as.factor(dens$intervention),
model = "equal",
xlab = "", ylab = "",
col = viridis::viridis(4, end = 0.8)[c(2, 4)],
lty = c(3,1), yaxt = "n",
bandcol = 'LightGray',
lwd = (c(2,2)))
##
## Test of equal densities: p-value = 0.08
legend("topright", levels(dens$intervention), col = viridis::viridis(4, end = 0.8)[c(2, 4)], lty=c(3,1), lwd=(c(2,2)))
# Create x-axis label. See https://stackoverflow.com/questions/11198767/how-to-annotate-across-or-between-plots-in-multi-plot-panels-in-r
par(mar = c(0,0,0,0))
plot(1, 1, type = "n", frame.plot = FALSE, axes = FALSE) # Fake plot for x-axis label
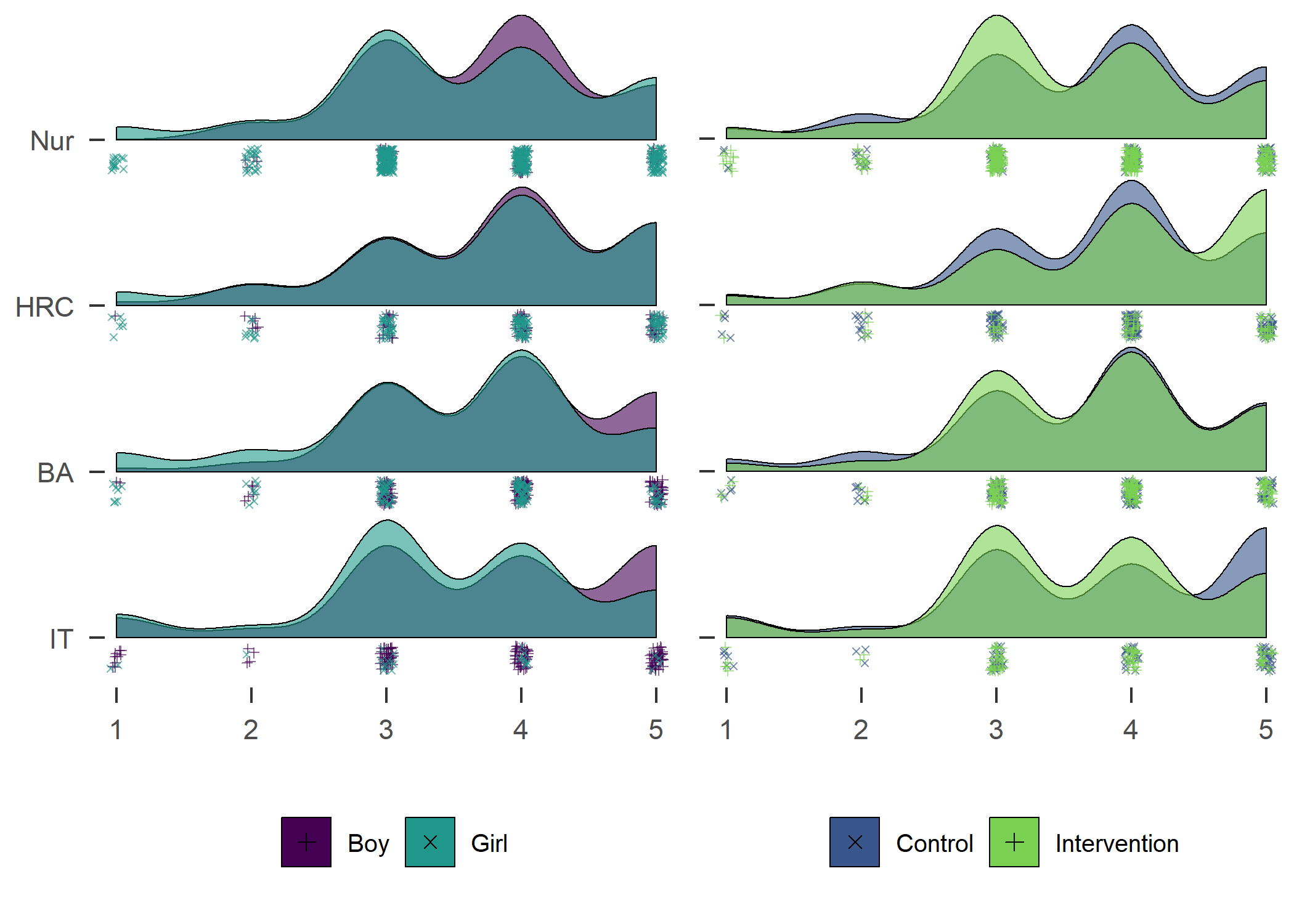
text(x = 1.02, y = 1.3, labels = "nordpaq_alternative_item hours", pos = 1)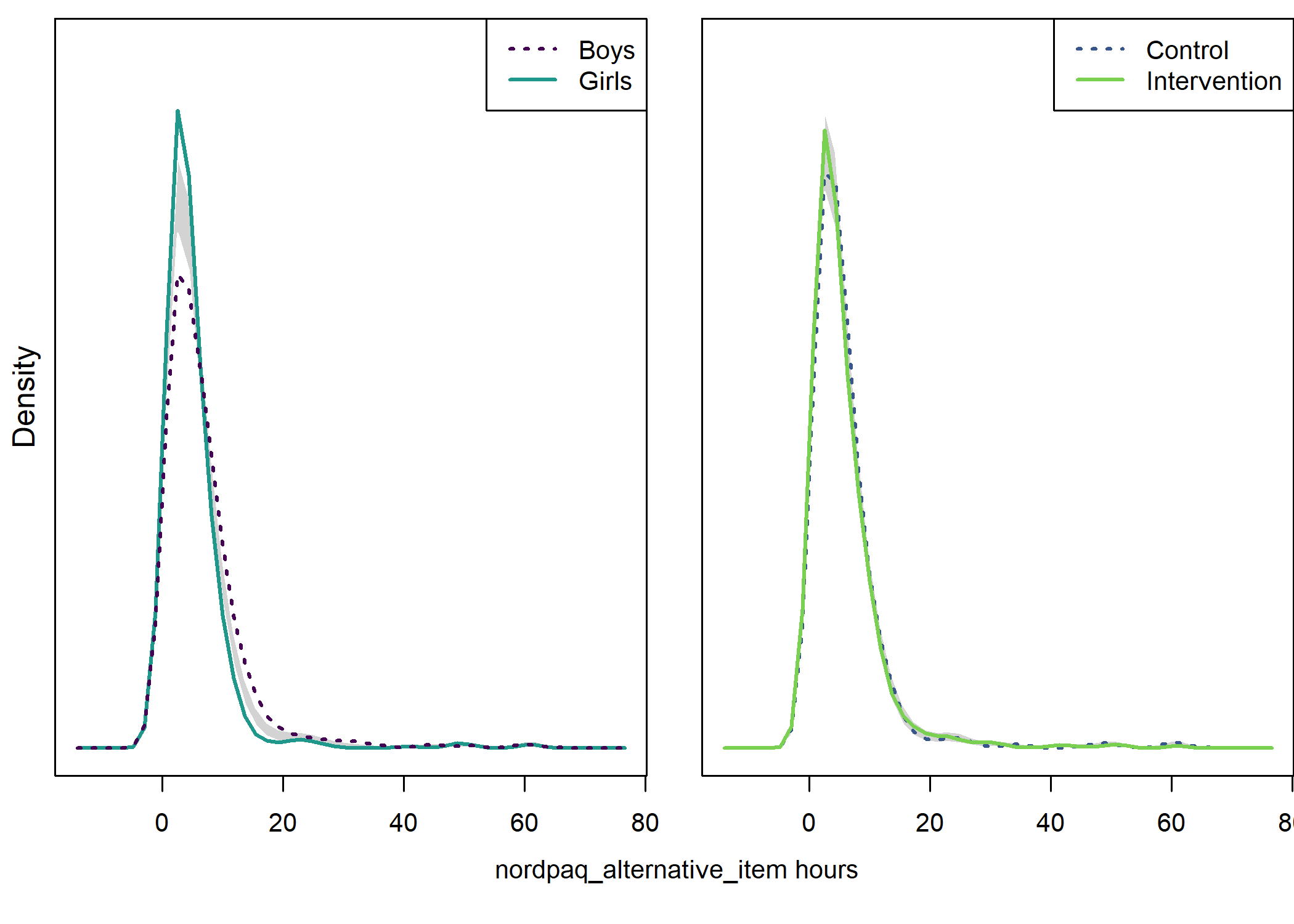
Data preparation
Description
The tab “Information on data preparation” of this section present information on data preparation for the plot
Information on data preparation
Prepare data
# m_nordpaq_alternative_itemAccelerometer_trackgirl <- brms::bf(leisuretimeMvpaHoursLastweek_T1 ~ (1 | track:girl)) %>%
# brms::brm(., data = df, chains = 4, iter = 4000, control = list(adapt_delta = 0.95))
#
# brms::prior_summary(m_nordpaq_alternative_itemAccelerometer_trackgirl, data = df)
#
# m_nordpaq_alternative_itemAccelerometer_trackgirl
#
# b_intercept <- brms::fixef(m_nordpaq_alternative_itemAccelerometer_trackgirl)[1]
#
# # This gives estimates only:
# nordpaq_alternative_item_trackgirl_estimates <- brms::ranef(m_nordpaq_alternative_itemAccelerometer_trackgirl)[[1]][1:10]
#
# # The 2.5%ile:
# nordpaq_alternative_item_trackgirl_CIL <- brms::ranef(m_nordpaq_alternative_itemAccelerometer_trackgirl)[[1]][21:30]
#
# # The 97.5%ile:
# nordpaq_alternative_item_trackgirl_CIH <- brms::ranef(m_nordpaq_alternative_itemAccelerometer_trackgirl)[[1]][31:40]
#
# nordpaq_alternative_item_trackgirl_ci <- data.frame(nordpaq_alternative_item_trackgirl_CIL, nordpaq_alternative_item_trackgirl_estimates, nordpaq_alternative_item_trackgirl_CIH, Variable = labels(brms::ranef(m_nordpaq_alternative_itemAccelerometer_trackgirl))) %>%
# dplyr::mutate(b_intercept = rep(b_intercept, 10)) %>%
# dplyr::mutate(nordpaq_alternative_item_trackgirl_CIL = nordpaq_alternative_item_trackgirl_CIL + b_intercept,
# nordpaq_alternative_item_trackgirl_estimates = nordpaq_alternative_item_trackgirl_estimates + b_intercept,
# nordpaq_alternative_item_trackgirl_CIH = nordpaq_alternative_item_trackgirl_CIH + b_intercept,
# track = c('BA_boy',
# 'BA_girl',
# 'HRC_boy',
# 'HRC_girl',
# 'IT_boy',
# 'IT_girl',
# 'Nur_boy',
# 'Nur_girl',
# 'Other_boy',
# 'Other_girl'
# ))
#
# nordpaq_alternative_item_trackgirl_diamonds_girl <- rbind(
# nordpaq_alternative_item_trackgirl_ci %>% dplyr::filter(track == "Nur_girl"),
# nordpaq_alternative_item_trackgirl_ci %>% dplyr::filter(track == "HRC_girl"),
# nordpaq_alternative_item_trackgirl_ci %>% dplyr::filter(track == "BA_girl"),
# nordpaq_alternative_item_trackgirl_ci %>% dplyr::filter(track == "IT_girl")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
# nordpaq_alternative_item_trackgirl_diamonds_boy <- rbind(
# nordpaq_alternative_item_trackgirl_ci %>% dplyr::filter(track == "Nur_boy"),
# nordpaq_alternative_item_trackgirl_ci %>% dplyr::filter(track == "HRC_boy"),
# nordpaq_alternative_item_trackgirl_ci %>% dplyr::filter(track == "BA_boy"),
# nordpaq_alternative_item_trackgirl_ci %>% dplyr::filter(track == "IT_boy")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
#
# m_nordpaq_alternative_itemAccelerometer_trackintervention <- brms::bf(leisuretimeMvpaHoursLastweek_T1 ~ (1 | track:intervention)) %>%
# brms::brm(., data = df, chains = 4, iter = 4000, control = list(adapt_delta = 0.95))
#
# brms::prior_summary(m_nordpaq_alternative_itemAccelerometer_trackintervention, data = df)
#
# m_nordpaq_alternative_itemAccelerometer_trackintervention
#
# b_intercept <- brms::fixef(m_nordpaq_alternative_itemAccelerometer_trackintervention)[1]
#
# # This gives estimates only:
# nordpaq_alternative_item_trackintervention_estimates <- brms::ranef(m_nordpaq_alternative_itemAccelerometer_trackintervention)[[1]][1:10]
#
#
# # The 2.5%ile:
# nordpaq_alternative_item_trackintervention_CIL <- brms::ranef(m_nordpaq_alternative_itemAccelerometer_trackintervention)[[1]][21:30]
#
# # The 97.5%ile:
# nordpaq_alternative_item_trackintervention_CIH <- brms::ranef(m_nordpaq_alternative_itemAccelerometer_trackintervention)[[1]][31:40]
#
# nordpaq_alternative_item_trackintervention_ci <- data.frame(nordpaq_alternative_item_trackintervention_CIL, nordpaq_alternative_item_trackintervention_estimates, nordpaq_alternative_item_trackintervention_CIH, Variable = labels(brms::ranef(m_nordpaq_alternative_itemAccelerometer_trackintervention))) %>%
# dplyr::mutate(b_intercept = rep(b_intercept, 10)) %>%
# dplyr::mutate(nordpaq_alternative_item_trackintervention_CIL = nordpaq_alternative_item_trackintervention_CIL + b_intercept,
# nordpaq_alternative_item_trackintervention_estimates = nordpaq_alternative_item_trackintervention_estimates + b_intercept,
# nordpaq_alternative_item_trackintervention_CIH = nordpaq_alternative_item_trackintervention_CIH + b_intercept,
# track = c('BA_control',
# 'BA_intervention',
# 'HRC_control',
# 'HRC_intervention',
# 'IT_control',
# 'IT_intervention',
# 'Nur_control',
# 'Nur_intervention',
# 'Other_control',
# 'Other_intervention'
# ))
#
# nordpaq_alternative_item_trackintervention_diamonds_intervention <- rbind(
# nordpaq_alternative_item_trackintervention_ci %>% dplyr::filter(track == "Nur_intervention"),
# nordpaq_alternative_item_trackintervention_ci %>% dplyr::filter(track == "HRC_intervention"),
# nordpaq_alternative_item_trackintervention_ci %>% dplyr::filter(track == "BA_intervention"),
# nordpaq_alternative_item_trackintervention_ci %>% dplyr::filter(track == "IT_intervention")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
# nordpaq_alternative_item_trackintervention_diamonds_control <- rbind(
# nordpaq_alternative_item_trackintervention_ci %>% dplyr::filter(track == "Nur_control"),
# nordpaq_alternative_item_trackintervention_ci %>% dplyr::filter(track == "HRC_control"),
# nordpaq_alternative_item_trackintervention_ci %>% dplyr::filter(track == "BA_control"),
# nordpaq_alternative_item_trackintervention_ci %>% dplyr::filter(track == "IT_control")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
# save(nordpaq_alternative_item_trackgirl_diamonds_girl, file = "./Rdata_files/nordpaq_alternative_item_trackgirl_diamonds_girl.Rdata")
# save(nordpaq_alternative_item_trackgirl_diamonds_boy, file = "./Rdata_files/nordpaq_alternative_item_trackgirl_diamonds_boy.Rdata")
# save(nordpaq_alternative_item_trackintervention_diamonds_intervention, file = "./Rdata_files/nordpaq_alternative_item_trackintervention_diamonds_intervention.Rdata")
# save(nordpaq_alternative_item_trackintervention_diamonds_control, file = "./Rdata_files/nordpaq_alternative_item_trackintervention_diamonds_control.Rdata")
load("./Rdata_files/nordpaq_alternative_item_trackgirl_diamonds_girl.Rdata")
load("./Rdata_files/nordpaq_alternative_item_trackgirl_diamonds_boy.Rdata")
load("./Rdata_files/nordpaq_alternative_item_trackintervention_diamonds_intervention.Rdata")
load("./Rdata_files/nordpaq_alternative_item_trackintervention_diamonds_control.Rdata")
nordpaq_alternative_item_trackgirl_diamonds_girlSensitivity analyses and robustness checks
Ideally, one would perform sensitivity analyses and robustness checks; e.g. comparing the estimates with frequentist multilevel models and following the WAMBS-checklist. Due to resource constraints, we are forced to forego this phase of analysis and instead, only show the linear model results for intercepts in each intervention-gender-track combination separately.
# To test the code with a single variable:
df_for_models_nested <- df %>%
dplyr::select(track, leisuretimeMvpaHoursLastweek_T1, girl, intervention) %>%
dplyr::mutate(track = forcats::fct_recode(track, NULL = "Other")) %>%
na.omit() %>%
dplyr::group_by(girl, intervention, track) %>%
tidyr::nest()
# This produced a data frame with columns "girl", "intervention", "track" and "data", the last of which is a data frame in each cell. E.g. for row wit intervention group girls in IT, there's a data frame for their values in the last column.
df_fitted <- df_for_models_nested %>%
dplyr::mutate(fit = map(data, ~ lm(leisuretimeMvpaHoursLastweek_T1 ~ 1, data = .x)))
# Now there's a linear model for each combination in the data frame
df_fitted <- df_fitted %>%
dplyr::mutate(
mean = map_dbl(fit, ~ coef(.x)[["(Intercept)"]]),
ci_low = map_dbl(fit, ~ confint(.x)["(Intercept)", 1]),
ci_high = map_dbl(fit, ~ confint(.x)["(Intercept)", 2]),
nonmissings = map_dbl(fit, ~ nobs(.x))
) %>%
dplyr::mutate(mean = round(mean, 2),
ci_low = round(ci_low, 2),
ci_high = round(ci_high, 2))
## (here was an earlier attempt for random effects)
# df_fitted <- df_fitted %>%
# dplyr::mutate(
# mean = map_dbl(fit, ~ lme4::fixef(.x)),
# m_p = map(fit, ~ profile(.x)),
# ci_low = map_dbl(m_p, ~ confint(.x)["(Intercept)", 1]),
# ci_high = map_dbl(m_p, ~ confint(.x)["(Intercept)", 2]),
# nonmissings = map_dbl(fit, ~ length(.x@resp$y))
# )
## (here was was an earlier attempt to use sandwich estimator
# df_for_sandwich_nested_withClusters <- df_for_sandwich_nested %>%
# dplyr::mutate(sandwich_clusters = purrr::map(data, magrittr::extract, c("group", "school", "track")))
# The last line selected the cluster variables from the data frame of each intervention-gender combination)
DT::datatable(df_fitted)Density plot of girls and boys in different educational tracks and schools
df %>% dplyr::select(leisuretimeMvpaHoursLastweek_T1, girl, trackSchool) %>%
group_by(girl, trackSchool) %>%
summarise(mean = mean(leisuretimeMvpaHoursLastweek_T1, na.rm = TRUE),
median = median(leisuretimeMvpaHoursLastweek_T1, na.rm = TRUE),
max = max(leisuretimeMvpaHoursLastweek_T1, na.rm = TRUE),
min = min(leisuretimeMvpaHoursLastweek_T1, na.rm = TRUE),
sd = sd(leisuretimeMvpaHoursLastweek_T1, na.rm = TRUE),
n = n()) %>%
DT::datatable() #papaja::apa_table()
df %>% dplyr::select(leisuretimeMvpaHoursLastweek_T1, girl, trackSchool) %>%
group_by(girl) %>%
summarise(mean = mean(leisuretimeMvpaHoursLastweek_T1, na.rm = TRUE),
median = median(leisuretimeMvpaHoursLastweek_T1, na.rm = TRUE),
max = max(leisuretimeMvpaHoursLastweek_T1, na.rm = TRUE),
min = min(leisuretimeMvpaHoursLastweek_T1, na.rm = TRUE),
sd = sd(leisuretimeMvpaHoursLastweek_T1, na.rm = TRUE),
n = n()) %>%
DT::datatable() #papaja::apa_table()
plot1 <- df %>% dplyr::select(id,
trackSchool = trackSchool,
girl,
nordpaq_alternative_item = leisuretimeMvpaHoursLastweek_T1) %>%
dplyr::filter(!is.na(trackSchool), !grepl('Other|NANA|BA4|HRC2|Nur2', trackSchool)) %>% # Drop categories with just few participants
ggplot2::ggplot(aes(y = trackSchool)) +
ggridges::geom_density_ridges2(aes(x = nordpaq_alternative_item, colour = "black",
fill = paste(trackSchool, girl)),
scale = 1,
alpha = 0.6, size = 0.25,
from = 0, # to = 450,
jittered_points=TRUE, point_shape=21,
point_fill="black") +
labs(x = "",
y = "") +
scale_y_discrete(expand = c(0.01, 0)) +
scale_x_continuous(expand = c(0.01, 0)) +
ggridges::scale_fill_cyclical(
labels = c('BA1 boy' = "Boy", 'BA1 girl' = "Girl"),
values = viridis::viridis(4, end = 0.8)[c(1, 3)],
name = "", guide = guide_legend(override.aes = list(alpha = 1))) +
ggridges::scale_colour_cyclical(values = "black") +
ggridges::theme_ridges(grid = FALSE) +
theme(legend.position="bottom", axis.text=element_text(size=10))
plot1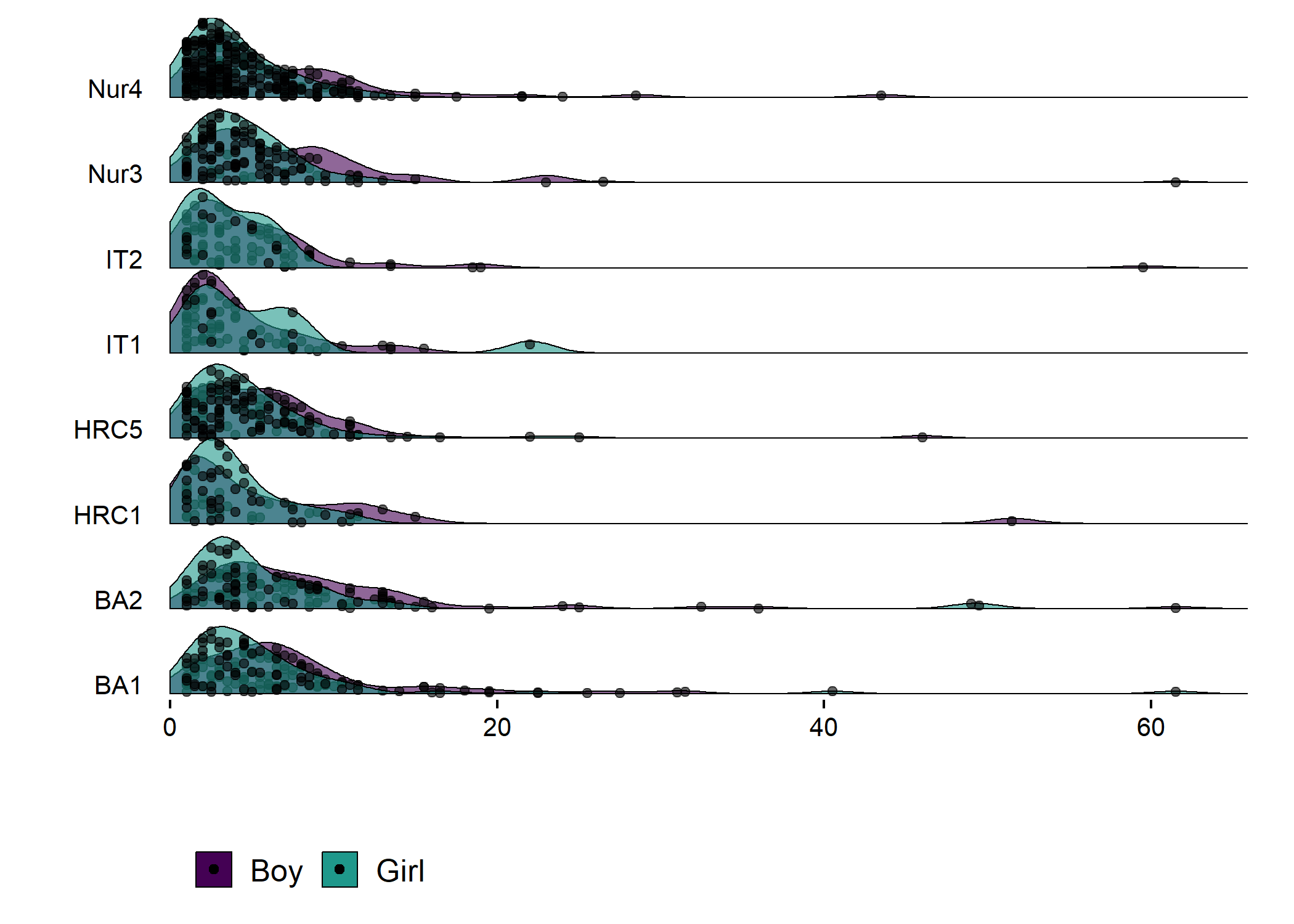
Density plot by track
plot1 <- df %>% dplyr::select(id,
track = track,
girl,
nordpaq_alternative_item = leisuretimeMvpaHoursLastweek_T1) %>%
dplyr::filter(!is.na(track), track != "Other") %>% # Drop category "Other"
dplyr::mutate(track = factor(track, levels = c("IT", "BA", "HRC", "Nur")),
track = recode_factor(track, "IT" = "IT", "BA" = "BA",
"HRC" = "HRC", "Nur" = "Nur")) %>%
ggplot2::ggplot(aes(y = track)) +
ggridges::geom_density_ridges2(aes(x = nordpaq_alternative_item, colour = "black",
fill = paste(track, girl),
point_color = girl,
point_fill = girl,
point_shape = girl),
scale = .75, alpha = 0.6, size = 0.25,
position = position_raincloud(width = 0.05, height = 0.15),
# from = 0, to = 450,
jittered_points = TRUE,
point_size = 1) +
labs(x = NULL,
y = NULL) +
scale_y_discrete(expand = c(0.1, 0)) +
scale_x_continuous(expand = c(0.01, 0)) +
ggridges::scale_fill_cyclical(
labels = c('BA boy' = "Boy", 'BA girl' = "Girl"),
values = viridis::viridis(4, end = 0.8)[c(1, 3)],
name = "",
guide = guide_legend(override.aes = list(alpha = 1,
point_shape = c(3, 4),
point_size = 2))) +
ggridges::scale_colour_cyclical(values = "black") +
ggridges::theme_ridges(grid = FALSE) +
# ggplot2::scale_fill_manual(values = c("#A0FFA0", "#A0A0FF")) +
ggridges::scale_discrete_manual(aesthetics = "point_color",
values = viridis::viridis(4, end = 0.8)[c(3, 1)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_fill",
values = viridis::viridis(4, end = 0.8)[c(3, 1)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_shape",
values = c(4, 3),
guide = "none") +
papaja::theme_apa() +
theme(legend.position = "bottom")
plot1 <- plot1 +
userfriendlyscience::diamondPlot(nordpaq_alternative_item_trackgirl_diamonds_girl,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[3],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15)) + # specify y-position; option to move the diamond
userfriendlyscience::diamondPlot(nordpaq_alternative_item_trackgirl_diamonds_boy,
returnLayerOnly = TRUE,
lineColor = "black",
linetype = "solid",
color=viridis::viridis(4, end = 0.8)[1],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15))
# Draw plot with intervention and control densities
plot2 <- df %>% dplyr::select(id,
track = track,
intervention,
nordpaq_alternative_item = leisuretimeMvpaHoursLastweek_T1) %>%
dplyr::filter(!is.na(track), track != "Other") %>% # Drop category "Other"
dplyr::mutate(track = factor(track, levels = c("IT", "BA", "HRC", "Nur")),
track = recode_factor(track, "IT" = "IT", "BA" = "BA",
"HRC" = "HRC", "Nur" = "Nur")) %>%
ggplot2::ggplot(aes(y = track)) +
ggridges::geom_density_ridges2(aes(x = nordpaq_alternative_item, colour = "black",
fill = paste(track, intervention),
point_color = intervention,
point_fill = intervention,
point_shape = intervention),
scale = .75, alpha = 0.6, size = 0.25,
position = position_raincloud(width = 0.05, height = 0.15),
# from = 0, to = 450,
jittered_points = TRUE,
point_size = 1) +
labs(x = NULL,
y = NULL) +
scale_y_discrete(expand = c(0.1, 0), labels = NULL) +
scale_x_continuous(expand = c(0.01, 0)) +
ggridges::scale_fill_cyclical(
labels = c('BA 0' = "Control", 'BA 1' = "Intervention"),
values = viridis::viridis(4, end = 0.8)[c(2, 4)],
name = "",
guide = guide_legend(override.aes = list(alpha = 1,
point_shape = c(4, 3),
point_size = 2))) +
ggridges::scale_colour_cyclical(values = "black") +
ggridges::theme_ridges(grid = FALSE) +
# ggplot2::scale_fill_manual(values = c("#A0FFA0", "#A0A0FF")) +
ggridges::scale_discrete_manual(aesthetics = "point_color",
values = viridis::viridis(4, end = 0.8)[c(2, 4)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_fill",
values = viridis::viridis(4, end = 0.8)[c(2, 4)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_shape",
values = c(4, 3),
guide = "none") +
papaja::theme_apa() +
theme(legend.position = "bottom")
plot2 <- plot2 +
userfriendlyscience::diamondPlot(nordpaq_alternative_item_trackintervention_diamonds_intervention,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[4],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15)) +
userfriendlyscience::diamondPlot(nordpaq_alternative_item_trackintervention_diamonds_control,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[2],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15))
plot1 + plot2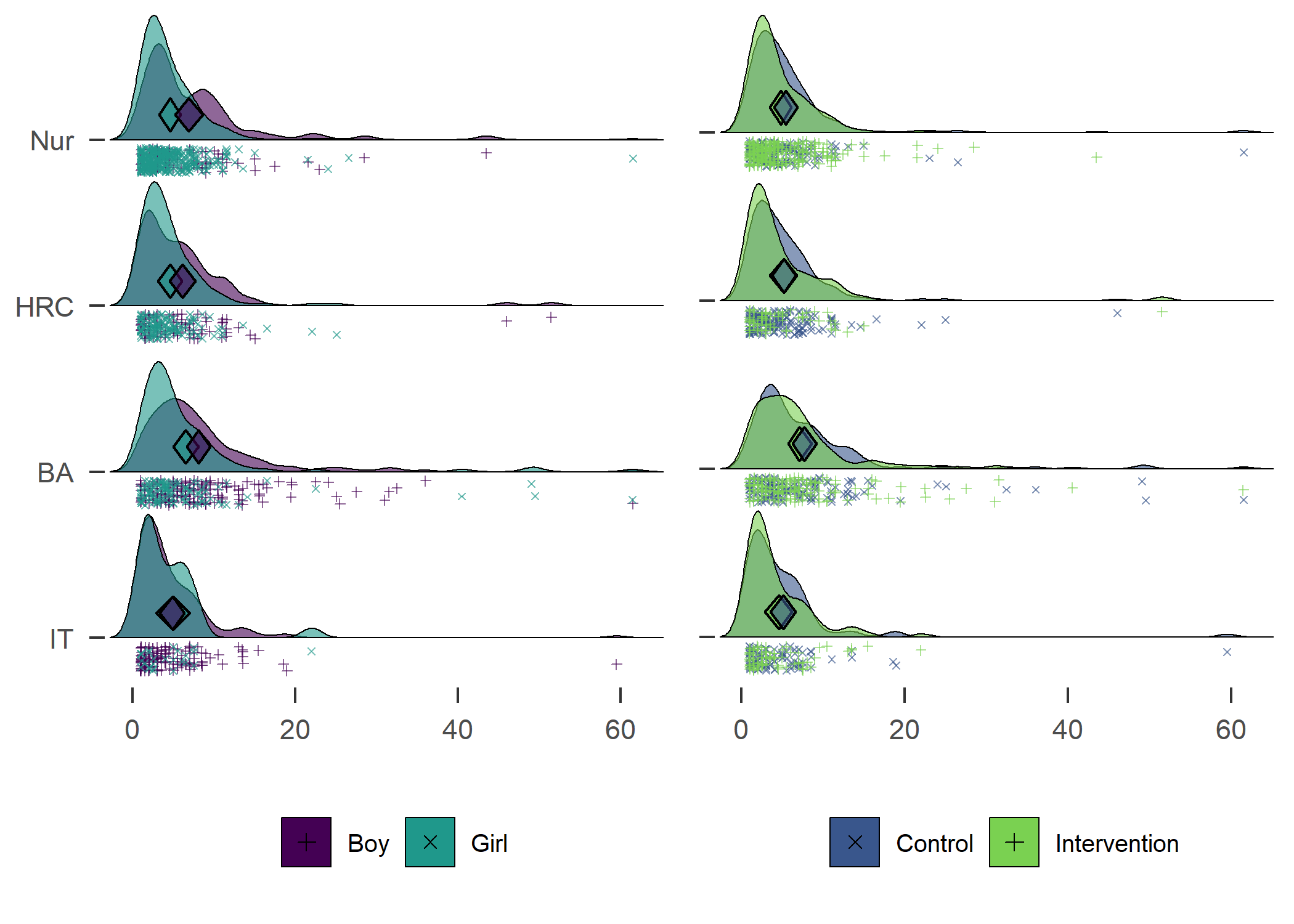
Self-reported minutes of MVPA during the previous week. Nur = Practical nurse, HRC = Hotel, restaurant and catering, BA = Business and administration, IT = Information and communications technology.
Breaks in sedentary time
This section reports the accelerometer-measured number of breaks to sitting time.
Data preparation
Description
The tab “Information on data preparation” of this section present information on data preparation for the plot
Information on data preparation
Code chunk below prepares data.
# m_sitBreaksAccelerometer_trackgirl <- brms::bf(sitBreaksAccelerometer_T1 ~ (1 | track:girl)) %>%
# brms::brm(., data = df, chains = 4, iter = 4000, control = list(adapt_delta = 0.95))
#
# brms::prior_summary(m_sitBreaksAccelerometer_trackgirl, data = df)
#
# m_sitBreaksAccelerometer_trackgirl
#
# b_intercept <- brms::fixef(m_sitBreaksAccelerometer_trackgirl)[1]
#
# # This gives estimates only:
# sitBreaksAccelerometer_trackgirl_estimates <- brms::ranef(m_sitBreaksAccelerometer_trackgirl)[[1]][1:10]
#
# # The 2.5%ile:
# sitBreaksAccelerometer_trackgirl_CIL <- brms::ranef(m_sitBreaksAccelerometer_trackgirl)[[1]][21:30]
#
# # The 97.5%ile:
# sitBreaksAccelerometer_trackgirl_CIH <- brms::ranef(m_sitBreaksAccelerometer_trackgirl)[[1]][31:40]
#
# sitBreaksAccelerometer_trackgirl_ci <- data.frame(sitBreaksAccelerometer_trackgirl_CIL, sitBreaksAccelerometer_trackgirl_estimates, sitBreaksAccelerometer_trackgirl_CIH, Variable = labels(brms::ranef(m_sitBreaksAccelerometer_trackgirl))) %>%
# dplyr::mutate(b_intercept = rep(b_intercept, 10)) %>%
# dplyr::mutate(sitBreaksAccelerometer_trackgirl_CIL = sitBreaksAccelerometer_trackgirl_CIL + b_intercept,
# sitBreaksAccelerometer_trackgirl_estimates = sitBreaksAccelerometer_trackgirl_estimates + b_intercept,
# sitBreaksAccelerometer_trackgirl_CIH = sitBreaksAccelerometer_trackgirl_CIH + b_intercept,
# track = c('BA_boy',
# 'BA_girl',
# 'HRC_boy',
# 'HRC_girl',
# 'IT_boy',
# 'IT_girl',
# 'Nur_boy',
# 'Nur_girl',
# 'Other_boy',
# 'Other_girl'
# ))
#
# sitBreaksAccelerometer_trackgirl_diamonds_girl <- rbind(
# sitBreaksAccelerometer_trackgirl_ci %>% dplyr::filter(track == "Nur_girl"),
# sitBreaksAccelerometer_trackgirl_ci %>% dplyr::filter(track == "HRC_girl"),
# sitBreaksAccelerometer_trackgirl_ci %>% dplyr::filter(track == "BA_girl"),
# sitBreaksAccelerometer_trackgirl_ci %>% dplyr::filter(track == "IT_girl")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
# sitBreaksAccelerometer_trackgirl_diamonds_boy <- rbind(
# sitBreaksAccelerometer_trackgirl_ci %>% dplyr::filter(track == "Nur_boy"),
# sitBreaksAccelerometer_trackgirl_ci %>% dplyr::filter(track == "HRC_boy"),
# sitBreaksAccelerometer_trackgirl_ci %>% dplyr::filter(track == "BA_boy"),
# sitBreaksAccelerometer_trackgirl_ci %>% dplyr::filter(track == "IT_boy")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
#
# m_sitBreaksAccelerometer_trackintervention <- brms::bf(sitBreaksAccelerometer_T1 ~ (1 | track:intervention)) %>%
# brms::brm(., data = df, chains = 4, iter = 4000, control = list(adapt_delta = 0.95))
#
# brms::prior_summary(m_sitBreaksAccelerometer_trackintervention, data = df)
#
# m_sitBreaksAccelerometer_trackintervention
#
# b_intercept <- brms::fixef(m_sitBreaksAccelerometer_trackintervention)[1]
#
# # This gives estimates only:
# sitBreaksAccelerometer_trackintervention_estimates <- brms::ranef(m_sitBreaksAccelerometer_trackintervention)[[1]][1:10]
#
# # The 2.5%ile:
# sitBreaksAccelerometer_trackintervention_CIL <- brms::ranef(m_sitBreaksAccelerometer_trackintervention)[[1]][21:30]
#
# # The 97.5%ile:
# sitBreaksAccelerometer_trackintervention_CIH <- brms::ranef(m_sitBreaksAccelerometer_trackintervention)[[1]][31:40]
#
# sitBreaksAccelerometer_trackintervention_ci <- data.frame(sitBreaksAccelerometer_trackintervention_CIL, sitBreaksAccelerometer_trackintervention_estimates, sitBreaksAccelerometer_trackintervention_CIH, Variable = labels(brms::ranef(m_sitBreaksAccelerometer_trackintervention))) %>%
# dplyr::mutate(b_intercept = rep(b_intercept, 10)) %>%
# dplyr::mutate(sitBreaksAccelerometer_trackintervention_CIL = sitBreaksAccelerometer_trackintervention_CIL + b_intercept,
# sitBreaksAccelerometer_trackintervention_estimates = sitBreaksAccelerometer_trackintervention_estimates + b_intercept,
# sitBreaksAccelerometer_trackintervention_CIH = sitBreaksAccelerometer_trackintervention_CIH + b_intercept,
# track = c('BA_control',
# 'BA_intervention',
# 'HRC_control',
# 'HRC_intervention',
# 'IT_control',
# 'IT_intervention',
# 'Nur_control',
# 'Nur_intervention',
# 'Other_control',
# 'Other_intervention'
# ))
#
# sitBreaksAccelerometer_trackintervention_diamonds_intervention <- rbind(
# sitBreaksAccelerometer_trackintervention_ci %>% dplyr::filter(track == "Nur_intervention"),
# sitBreaksAccelerometer_trackintervention_ci %>% dplyr::filter(track == "HRC_intervention"),
# sitBreaksAccelerometer_trackintervention_ci %>% dplyr::filter(track == "BA_intervention"),
# sitBreaksAccelerometer_trackintervention_ci %>% dplyr::filter(track == "IT_intervention")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
# sitBreaksAccelerometer_trackintervention_diamonds_control <- rbind(
# sitBreaksAccelerometer_trackintervention_ci %>% dplyr::filter(track == "Nur_control"),
# sitBreaksAccelerometer_trackintervention_ci %>% dplyr::filter(track == "HRC_control"),
# sitBreaksAccelerometer_trackintervention_ci %>% dplyr::filter(track == "BA_control"),
# sitBreaksAccelerometer_trackintervention_ci %>% dplyr::filter(track == "IT_control")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
# save(sitBreaksAccelerometer_trackgirl_diamonds_girl, file = "./Rdata_files/sitBreaksAccelerometer_trackgirl_diamonds_girl.Rdata")
# save(sitBreaksAccelerometer_trackgirl_diamonds_boy, file = "./Rdata_files/sitBreaksAccelerometer_trackgirl_diamonds_boy.Rdata")
# save(sitBreaksAccelerometer_trackintervention_diamonds_intervention, file = "./Rdata_files/sitBreaksAccelerometer_trackintervention_diamonds_intervention.Rdata")
# save(sitBreaksAccelerometer_trackintervention_diamonds_control, file = "./Rdata_files/sitBreaksAccelerometer_trackintervention_diamonds_control.Rdata")
load("./Rdata_files/sitBreaksAccelerometer_trackgirl_diamonds_girl.Rdata")
load("./Rdata_files/sitBreaksAccelerometer_trackgirl_diamonds_boy.Rdata")
load("./Rdata_files/sitBreaksAccelerometer_trackintervention_diamonds_intervention.Rdata")
load("./Rdata_files/sitBreaksAccelerometer_trackintervention_diamonds_control.Rdata")Density plot by intervention and gender
# Create data frame
densplot <- d
levels(densplot$intervention) <- c("Control", "Intervention")
levels(densplot$girl) <- recode(densplot$girl, "boy" = "Boys", "girl" = "Girls")
# This gives side-by-side plots. There's a third plot below the two, which is fake to include x-axis text.
layout(matrix(c(1, 2, 3, 3), nrow = 2, ncol = 2, byrow = TRUE),
widths = c(0.5, 0.5), heights = c(0.45, 0.05))
# Minimise whitespace; see https://stackoverflow.com/questions/15848942/how-to-reduce-space-gap-between-multiple-graphs-in-r
par(mai = c(0.3, 0.3, 0.1, 0.0))
## Girls vs. boys
# Choose only the variables needed and drop NA
dens <- densplot %>% dplyr::select(sitBreaksAccelerometer_T1, girl) %>%
dplyr::filter(complete.cases(.))
# Set random number generator for reproducibility of bootstrap test of equal densities
set.seed(10)
# Make plot
sm.sitBreaksAccelerometer_T1_2 <- sm.density.compare2(as.numeric(dens$sitBreaksAccelerometer_T1),
as.factor(dens$girl),
model = "equal",
xlab = "", ylab = "",
col = viridis::viridis(4, end = 0.8)[c(3, 1)],
lty = c(1, 3), yaxt = "n",
bandcol = 'LightGray',
lwd = c(2, 2))
##
## Test of equal densities: p-value = 0
legend("topright", levels(dens$girl), col = viridis::viridis(4, end = 0.8)[c(1, 3)], lty = c(3,1), lwd = (c(2,2)))
mtext(side = 2, "Density", line = 0.5)
## Intervention vs. control
# Choose only the variables needed and drop NA
dens <- densplot %>% dplyr::select(sitBreaksAccelerometer_T1, intervention) %>%
dplyr::filter(complete.cases(.))
# Set random number generator for reproducibility of bootstrap test of equal densities
set.seed(10)
# Make plot
sm.sitBreaksAccelerometer_T1_2 <- sm.density.compare2(as.numeric(dens$sitBreaksAccelerometer_T1),
as.factor(dens$intervention),
model = "equal",
xlab = "", ylab = "",
col = viridis::viridis(4, end = 0.8)[c(2, 4)],
lty = c(3, 1), yaxt = "n",
bandcol = 'LightGray',
lwd = c(2, 2))
##
## Test of equal densities: p-value = 0.01
legend("topright", levels(dens$intervention), col = viridis::viridis(4, end = 0.8)[c(2, 4)], lty=c(3,1), lwd=(c(2,2)))
# Create x-axis label. See https://stackoverflow.com/questions/11198767/how-to-annotate-across-or-between-plots-in-multi-plot-panels-in-r
par(mar = c(0,0,0,0))
plot(1, 1, type = "n", frame.plot = FALSE, axes = FALSE) # Fake plot for x-axis label
text(x = 1.02, y = 1.3, labels = "Average number of breaks in daily sitting", pos = 1)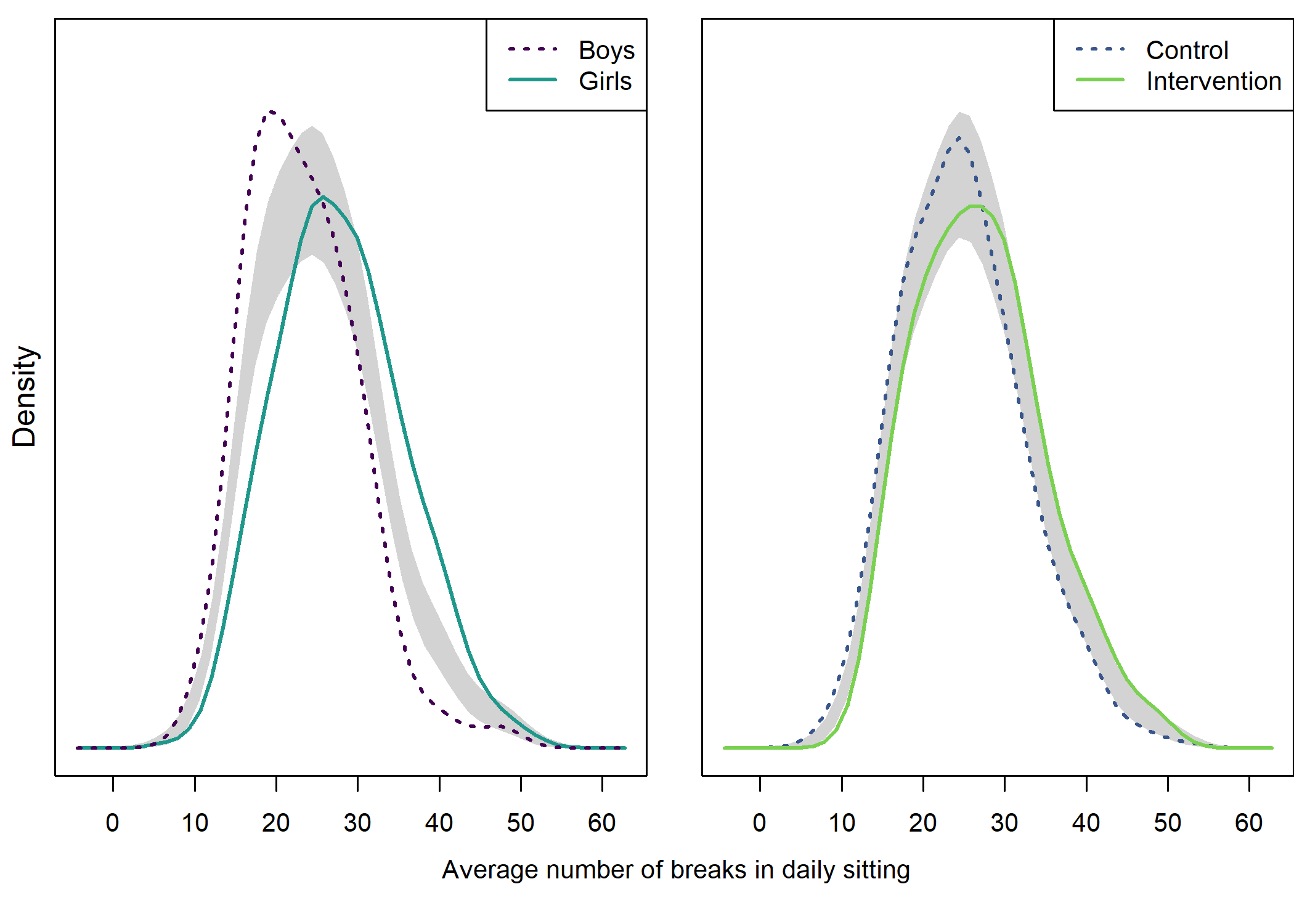
Density plot by track
plot1 <- df %>% dplyr::select(id,
track = track,
girl,
PA = sitBreaksAccelerometer_T1) %>%
dplyr::filter(!is.na(track), track != "Other") %>% # Drop category "Other"
dplyr::mutate(track = factor(track, levels = c("IT", "BA", "HRC", "Nur")),
track = recode_factor(track, "IT" = "IT", "BA" = "BA",
"HRC" = "HRC", "Nur" = "Nur")) %>%
ggplot2::ggplot(aes(y = track)) +
ggridges::geom_density_ridges2(aes(x = PA, colour = "black",
fill = paste(track, girl),
point_color = girl,
point_fill = girl,
point_shape = girl),
scale = .6, alpha = 0.6, size = 0.25,
position = position_raincloud(width = 0.2, height = 0.15),
# from = 0, to = 450,
jittered_points = TRUE,
point_size = 1) +
labs(x = NULL,
y = NULL) +
scale_y_discrete(expand = c(0.1, 0)) +
scale_x_continuous(expand = c(0.01, 0)) +
ggridges::scale_fill_cyclical(
labels = c('BA boy' = "Boy", 'BA girl' = "Girl"),
values = viridis::viridis(4, end = 0.8)[c(1, 3)],
name = "",
guide = guide_legend(override.aes = list(alpha = 1,
point_shape = c(3, 4),
point_size = 2))) +
ggridges::scale_colour_cyclical(values = "black") +
ggridges::theme_ridges(grid = FALSE) +
# ggplot2::scale_fill_manual(values = c("#A0FFA0", "#A0A0FF")) +
ggridges::scale_discrete_manual(aesthetics = "point_color",
values = viridis::viridis(4, end = 0.8)[c(3, 1)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_fill",
values = viridis::viridis(4, end = 0.8)[c(3, 1)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_shape",
values = c(4, 3),
guide = "none") +
papaja::theme_apa() +
theme(legend.position = "bottom")
plot1 <- plot1 +
userfriendlyscience::diamondPlot(sitBreaksAccelerometer_trackgirl_diamonds_girl,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[3],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15)) + # specify y-position; option to move the diamond
userfriendlyscience::diamondPlot(sitBreaksAccelerometer_trackgirl_diamonds_boy,
returnLayerOnly = TRUE,
lineColor = "black",
linetype = "solid",
color=viridis::viridis(4, end = 0.8)[1],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15))
# Draw plot with intervention and control densities
plot2 <- df %>% dplyr::select(id,
track = track,
intervention,
PA = sitBreaksAccelerometer_T1) %>%
dplyr::filter(!is.na(track), track != "Other") %>% # Drop category "Other"
dplyr::mutate(track = factor(track, levels = c("IT", "BA", "HRC", "Nur")),
track = recode_factor(track, "IT" = "IT", "BA" = "BA",
"HRC" = "HRC", "Nur" = "Nur")) %>%
ggplot2::ggplot(aes(y = track)) +
ggridges::geom_density_ridges2(aes(x = PA, colour = "black",
fill = paste(track, intervention),
point_color = intervention,
point_fill = intervention,
point_shape = intervention),
scale = .6, alpha = 0.6, size = 0.25,
position = position_raincloud(width = 0.2, height = 0.15),
# from = 0, to = 450,
jittered_points = TRUE,
point_size = 1) +
labs(x = NULL,
y = NULL) +
scale_y_discrete(expand = c(0.1, 0), labels = NULL) +
scale_x_continuous(expand = c(0.01, 0)) +
ggridges::scale_fill_cyclical(
labels = c('BA 0' = "Control", 'BA 1' = "Intervention"),
values = viridis::viridis(4, end = 0.8)[c(2, 4)],
name = "",
guide = guide_legend(override.aes = list(alpha = 1,
point_shape = c(4, 3),
point_size = 2))) +
ggridges::scale_colour_cyclical(values = "black") +
ggridges::theme_ridges(grid = FALSE) +
# ggplot2::scale_fill_manual(values = c("#A0FFA0", "#A0A0FF")) +
ggridges::scale_discrete_manual(aesthetics = "point_color",
values = viridis::viridis(4, end = 0.8)[c(2, 4)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_fill",
values = viridis::viridis(4, end = 0.8)[c(2, 4)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_shape",
values = c(4, 3),
guide = "none") +
papaja::theme_apa() +
theme(legend.position = "bottom")
plot2 <- plot2 +
userfriendlyscience::diamondPlot(sitBreaksAccelerometer_trackintervention_diamonds_intervention,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[4],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15)) +
userfriendlyscience::diamondPlot(sitBreaksAccelerometer_trackintervention_diamonds_control,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[2],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15))
plot1 + plot2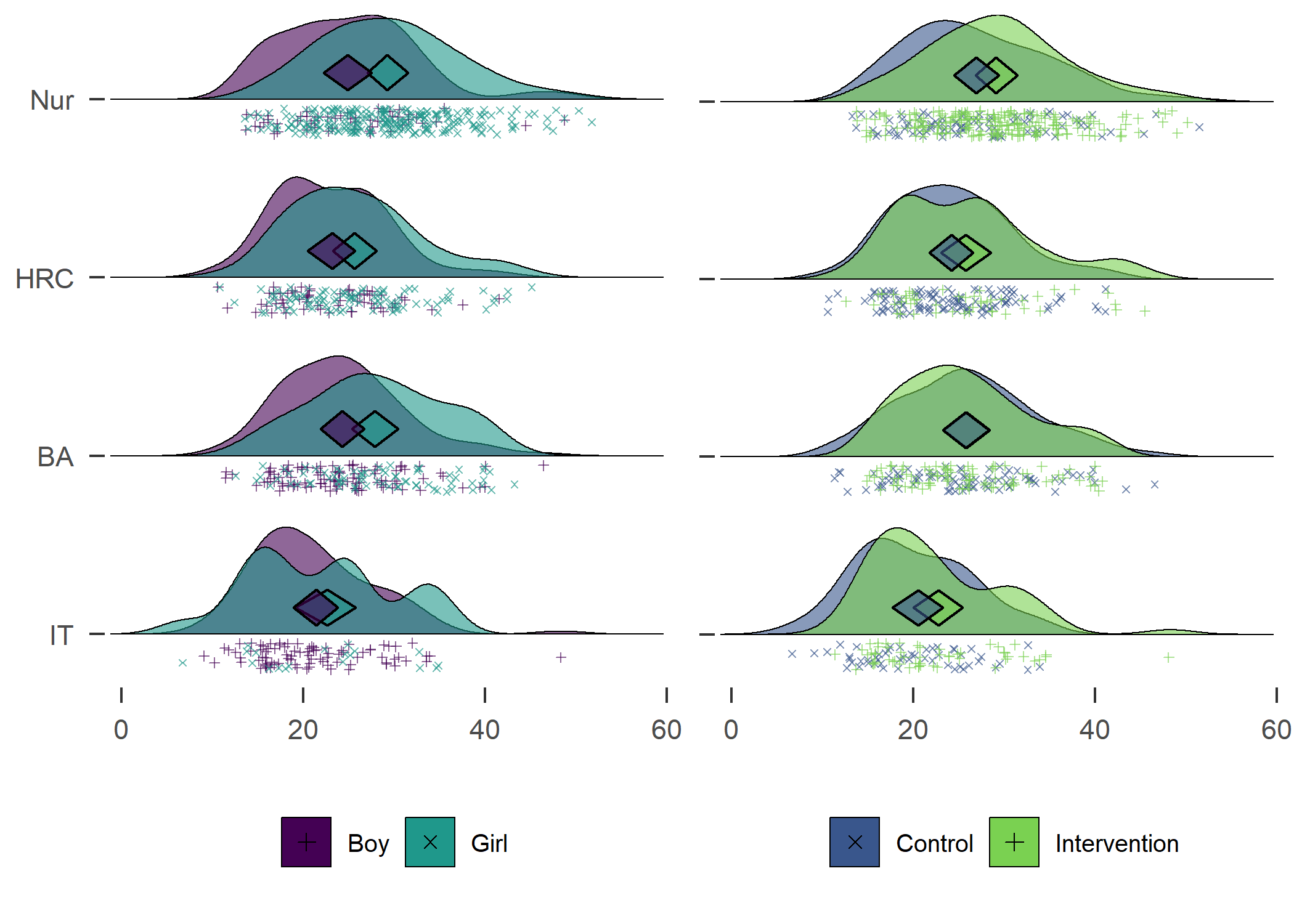
Breaks in sedentary time. Nur = Practical nurse, HRC = Hotel, restaurant and catering, BA = Business and administration, IT = Information and communications technology.
Girls in all educational tracks interrupted sitting more than boys.
SB, Accelerometer-measured
Density plot by intervention and gender
# Create data frame
densplot <- d
levels(densplot$intervention) <- c("Control", "Intervention")
levels(densplot$girl) <- recode(densplot$girl, "boy" = "Boys", "girl" = "Girls")
# This gives side-by-side plots. There's a third plot below the two, which is fake to include x-axis text.
layout(matrix(c(1, 2, 3, 3), nrow = 2, ncol = 2, byrow = TRUE),
widths = c(0.5, 0.5), heights = c(0.45, 0.05))
# Minimise whitespace; see https://stackoverflow.com/questions/15848942/how-to-reduce-space-gap-between-multiple-graphs-in-r
par(mai = c(0.3, 0.3, 0.1, 0.0))
## Girls vs. boys
# Choose only the variables needed and drop NA
dens <- densplot %>% dplyr::select(sitLieAccelerometer_T1, girl) %>%
dplyr::filter(complete.cases(.))
# Set random number generator for reproducibility of bootstrap test of equal densities
set.seed(10)
# Make plot
sm.sitLieAccelerometer_T1_2 <- sm.density.compare2(as.numeric(dens$sitLieAccelerometer_T1),
as.factor(dens$girl),
model = "equal",
xlab = "", ylab = "",
col = viridis::viridis(4, end = 0.8)[c(3, 1)],
lty = c(1, 3), yaxt = "n",
bandcol = 'LightGray',
lwd = c(2, 2))
##
## Test of equal densities: p-value = 0
legend("topright", levels(dens$girl), col = viridis::viridis(4, end = 0.8)[c(1, 3)], lty = c(3,1), lwd = (c(2,2)))
mtext(side = 2, "Density", line = 0.5)
## Intervention vs. control
# Choose only the variables needed and drop NA
dens <- densplot %>% dplyr::select(sitLieAccelerometer_T1, intervention) %>%
dplyr::filter(complete.cases(.))
# Set random number generator for reproducibility of bootstrap test of equal densities
set.seed(10)
# Make plot
sm.sitLieAccelerometer_T1_2 <- sm.density.compare2(as.numeric(dens$sitLieAccelerometer_T1),
as.factor(dens$intervention),
model = "equal",
xlab = "", ylab = "",
col = viridis::viridis(4, end = 0.8)[c(2, 4)],
lty = c(3, 1), yaxt = "n",
bandcol = 'LightGray',
lwd = c(2, 2))
##
## Test of equal densities: p-value = 0.07
legend("topright", levels(dens$intervention), col = viridis::viridis(4, end = 0.8)[c(2, 4)], lty=c(3,1), lwd=(c(2,2)))
# Create x-axis label. See https://stackoverflow.com/questions/11198767/how-to-annotate-across-or-between-plots-in-multi-plot-panels-in-r
par(mar = c(0,0,0,0))
plot(1, 1, type = "n", frame.plot = FALSE, axes = FALSE) # Fake plot for x-axis label
text(x = 1.02, y = 1.3, labels = "Average daily hours spent sitting or lying down", pos = 1)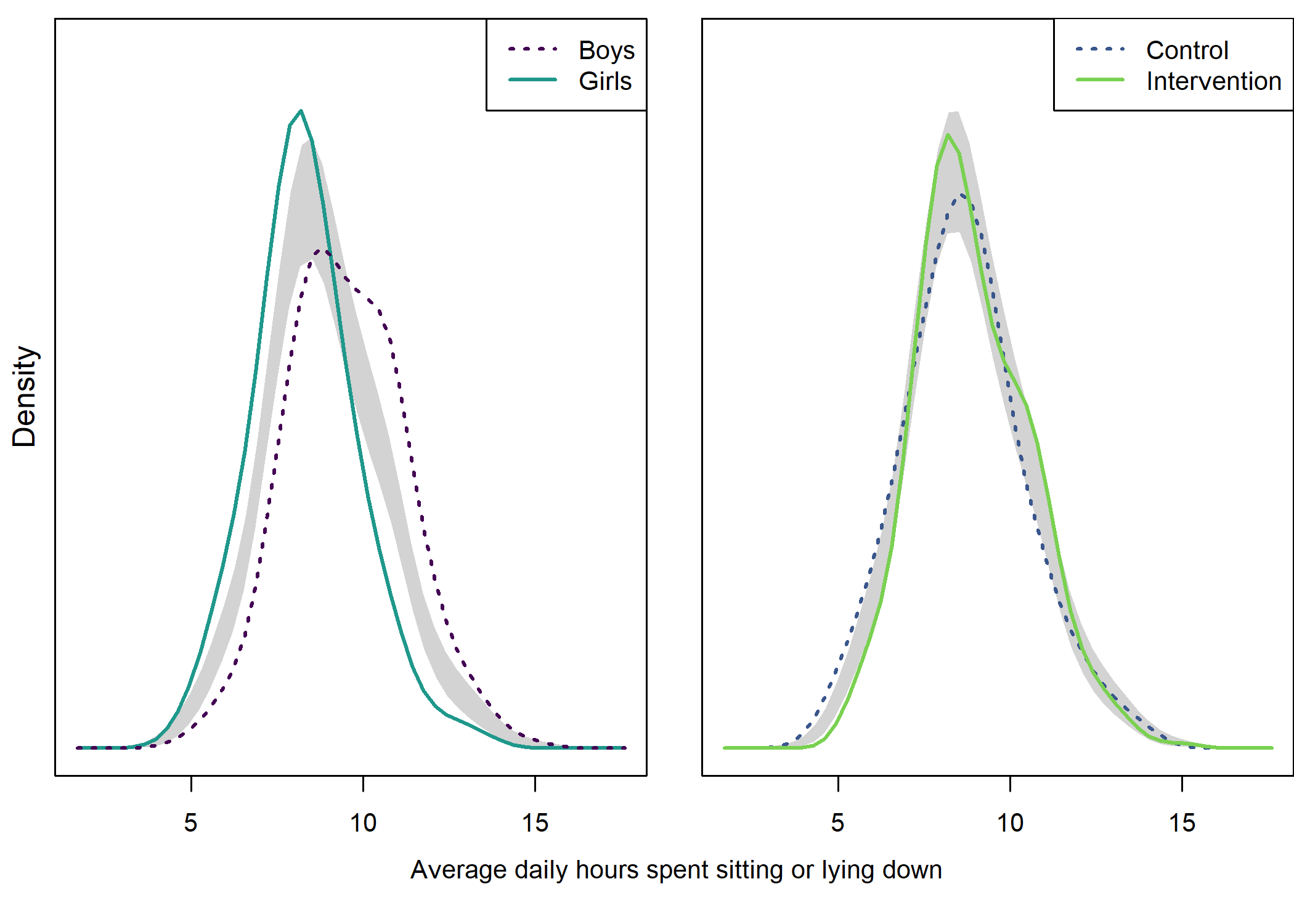
Differences in time spent sitting and lying down emerged between boys and girls, the former spending more time being sedentary. Even though there was some heterogeneity among schools, again the aggregated intervention and control schools showed no differences.
Data preparation
Description
The tab “Information on data preparation” of this section present information on data preparation for the plot
Information on data preparation
Prepare data: time spent sitting & lying down
Get credibility intervals track:school
# m_sitLieAccelerometer_trackgirl <- brms::bf(sitLieAccelerometer_T1 ~ (1 | track:girl)) %>%
# brms::brm(., data = df, chains = 4, iter = 4000, control = list(adapt_delta = 0.95))
#
# brms::prior_summary(m_sitLieAccelerometer_trackgirl, data = df)
#
# m_sitLieAccelerometer_trackgirl
#
# b_intercept <- brms::fixef(m_sitLieAccelerometer_trackgirl)[1]
#
# # This gives estimates only:
# sitLie_trackgirl_estimates <- brms::ranef(m_sitLieAccelerometer_trackgirl)[[1]][1:10]
#
# # The 2.5%ile:
# sitLie_trackgirl_CIL <- brms::ranef(m_sitLieAccelerometer_trackgirl)[[1]][21:30]
#
# # The 97.5%ile:
# sitLie_trackgirl_CIH <- brms::ranef(m_sitLieAccelerometer_trackgirl)[[1]][31:40]
#
# sitLie_trackgirl_ci <- data.frame(sitLie_trackgirl_CIL, sitLie_trackgirl_estimates, sitLie_trackgirl_CIH, Variable = labels(brms::ranef(m_sitLieAccelerometer_trackgirl))) %>%
# dplyr::mutate(b_intercept = rep(b_intercept, 10)) %>%
# dplyr::mutate(sitLie_trackgirl_CIL = sitLie_trackgirl_CIL + b_intercept,
# sitLie_trackgirl_estimates = sitLie_trackgirl_estimates + b_intercept,
# sitLie_trackgirl_CIH = sitLie_trackgirl_CIH + b_intercept,
# track = c('BA_boy',
# 'BA_girl',
# 'HRC_boy',
# 'HRC_girl',
# 'IT_boy',
# 'IT_girl',
# 'Nur_boy',
# 'Nur_girl',
# 'Other_boy',
# 'Other_girl'
# ))
#
# sitLie_trackgirl_diamonds_girl <- rbind(
# sitLie_trackgirl_ci %>% dplyr::filter(track == "Nur_girl"),
# sitLie_trackgirl_ci %>% dplyr::filter(track == "HRC_girl"),
# sitLie_trackgirl_ci %>% dplyr::filter(track == "BA_girl"),
# sitLie_trackgirl_ci %>% dplyr::filter(track == "IT_girl")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
# sitLie_trackgirl_diamonds_boy <- rbind(
# sitLie_trackgirl_ci %>% dplyr::filter(track == "Nur_boy"),
# sitLie_trackgirl_ci %>% dplyr::filter(track == "HRC_boy"),
# sitLie_trackgirl_ci %>% dplyr::filter(track == "BA_boy"),
# sitLie_trackgirl_ci %>% dplyr::filter(track == "IT_boy")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
#
# m_sitLieAccelerometer_trackintervention <- brms::bf(sitLieAccelerometer_T1 ~ (1 | track:intervention)) %>%
# brms::brm(., data = df, chains = 4, iter = 4000, control = list(adapt_delta = 0.95))
#
# brms::prior_summary(m_sitLieAccelerometer_trackintervention, data = df)
#
# m_sitLieAccelerometer_trackintervention
#
# b_intercept <- brms::fixef(m_sitLieAccelerometer_trackintervention)[1]
#
# # This gives estimates only:
# sitLie_trackintervention_estimates <- brms::ranef(m_sitLieAccelerometer_trackintervention)[[1]][1:10]
#
#
# # The 2.5%ile:
# sitLie_trackintervention_CIL <- brms::ranef(m_sitLieAccelerometer_trackintervention)[[1]][21:30]
#
# # The 97.5%ile:
# sitLie_trackintervention_CIH <- brms::ranef(m_sitLieAccelerometer_trackintervention)[[1]][31:40]
#
# sitLie_trackintervention_ci <- data.frame(sitLie_trackintervention_CIL, sitLie_trackintervention_estimates, sitLie_trackintervention_CIH, Variable = labels(brms::ranef(m_sitLieAccelerometer_trackintervention))) %>%
# dplyr::mutate(b_intercept = rep(b_intercept, 10)) %>%
# dplyr::mutate(sitLie_trackintervention_CIL = sitLie_trackintervention_CIL + b_intercept,
# sitLie_trackintervention_estimates = sitLie_trackintervention_estimates + b_intercept,
# sitLie_trackintervention_CIH = sitLie_trackintervention_CIH + b_intercept,
# track = c('BA_control',
# 'BA_intervention',
# 'HRC_control',
# 'HRC_intervention',
# 'IT_control',
# 'IT_intervention',
# 'Nur_control',
# 'Nur_intervention',
# 'Other_control',
# 'Other_intervention'
# ))
#
# sitLie_trackintervention_diamonds_intervention <- rbind(
# sitLie_trackintervention_ci %>% dplyr::filter(track == "Nur_intervention"),
# sitLie_trackintervention_ci %>% dplyr::filter(track == "HRC_intervention"),
# sitLie_trackintervention_ci %>% dplyr::filter(track == "BA_intervention"),
# sitLie_trackintervention_ci %>% dplyr::filter(track == "IT_intervention")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
# sitLie_trackintervention_diamonds_control <- rbind(
# sitLie_trackintervention_ci %>% dplyr::filter(track == "Nur_control"),
# sitLie_trackintervention_ci %>% dplyr::filter(track == "HRC_control"),
# sitLie_trackintervention_ci %>% dplyr::filter(track == "BA_control"),
# sitLie_trackintervention_ci %>% dplyr::filter(track == "IT_control")) %>%
# arrange(-row_number()) # reverse order of rows to make them right for the plot to come
#
# save(sitLie_trackgirl_diamonds_girl, file = "./Rdata_files/sitLie_trackgirl_diamonds_girl.Rdata")
# save(sitLie_trackgirl_diamonds_boy, file = "./Rdata_files/sitLie_trackgirl_diamonds_boy.Rdata")
# save(sitLie_trackintervention_diamonds_intervention, file = "./Rdata_files/sitLie_trackintervention_diamonds_intervention.Rdata")
# save(sitLie_trackintervention_diamonds_control, file = "./Rdata_files/sitLie_trackintervention_diamonds_control.Rdata")
load("./Rdata_files/sitLie_trackgirl_diamonds_girl.Rdata")
load("./Rdata_files/sitLie_trackgirl_diamonds_boy.Rdata")
load("./Rdata_files/sitLie_trackintervention_diamonds_intervention.Rdata")
load("./Rdata_files/sitLie_trackintervention_diamonds_control.Rdata")Basic observed characteristics
NOTE: These values do not account for clustering.
df %>% dplyr::select(sitLieAccelerometer_T1, girl, track) %>%
group_by(girl, track) %>%
summarise(mean = mean(sitLieAccelerometer_T1, na.rm = TRUE),
median = median(sitLieAccelerometer_T1, na.rm = TRUE),
max = max(sitLieAccelerometer_T1, na.rm = TRUE),
min = min(sitLieAccelerometer_T1, na.rm = TRUE),
sd = sd(sitLieAccelerometer_T1, na.rm = TRUE),
n = n()) %>%
DT::datatable() #papaja::apa_table()
df %>% dplyr::select(sitLieAccelerometer_T1, girl, track) %>%
group_by(girl) %>%
summarise(mean = mean(sitLieAccelerometer_T1, na.rm = TRUE),
median = median(sitLieAccelerometer_T1, na.rm = TRUE),
max = max(sitLieAccelerometer_T1, na.rm = TRUE),
min = min(sitLieAccelerometer_T1, na.rm = TRUE),
sd = sd(sitLieAccelerometer_T1, na.rm = TRUE),
n = n()) %>%
DT::datatable() #papaja::apa_table()
df %>% dplyr::select(sitLieAccelerometer_T1, intervention, track) %>%
group_by(intervention, track) %>%
summarise(mean = mean(sitLieAccelerometer_T1, na.rm = TRUE),
median = median(sitLieAccelerometer_T1, na.rm = TRUE),
max = max(sitLieAccelerometer_T1, na.rm = TRUE),
min = min(sitLieAccelerometer_T1, na.rm = TRUE),
sd = sd(sitLieAccelerometer_T1, na.rm = TRUE),
n = n()) %>%
DT::datatable() #papaja::apa_table()
df %>% dplyr::select(sitLieAccelerometer_T1, intervention, track) %>%
group_by(intervention) %>%
summarise(mean = mean(sitLieAccelerometer_T1, na.rm = TRUE),
median = median(sitLieAccelerometer_T1, na.rm = TRUE),
max = max(sitLieAccelerometer_T1, na.rm = TRUE),
min = min(sitLieAccelerometer_T1, na.rm = TRUE),
sd = sd(sitLieAccelerometer_T1, na.rm = TRUE),
n = n()) %>%
DT::datatable() #papaja::apa_table()Density plot by track
Time spent sitting or lying down
plot1 <- df %>% dplyr::select(id,
track = track,
girl,
PA = sitLieAccelerometer_T1) %>%
dplyr::filter(!is.na(track), track != "Other") %>% # Drop category "Other"
dplyr::mutate(track = factor(track, levels = c("IT", "BA", "HRC", "Nur")),
track = recode_factor(track, "IT" = "IT", "BA" = "BA",
"HRC" = "HRC", "Nur" = "Nur")) %>%
ggplot2::ggplot(aes(y = track)) +
ggridges::geom_density_ridges2(aes(x = PA, colour = "black",
fill = paste(track, girl),
point_color = girl,
point_fill = girl,
point_shape = girl),
scale = .75, alpha = 0.6, size = 0.25,
position = position_raincloud(width = 0.05, height = 0.15),
# from = 0, to = 450,
jittered_points = TRUE,
point_size = 1) +
labs(x = NULL,
y = NULL) +
scale_y_discrete(expand = c(0.1, 0)) +
scale_x_continuous(expand = c(0.01, 0)) +
ggridges::scale_fill_cyclical(
labels = c('BA boy' = "Boy", 'BA girl' = "Girl"),
values = viridis::viridis(4, end = 0.8)[c(1, 3)],
name = "",
guide = guide_legend(override.aes = list(alpha = 1,
point_shape = c(3, 4),
point_size = 2))) +
ggridges::scale_colour_cyclical(values = "black") +
ggridges::theme_ridges(grid = FALSE) +
# ggplot2::scale_fill_manual(values = c("#A0FFA0", "#A0A0FF")) +
ggridges::scale_discrete_manual(aesthetics = "point_color",
values = viridis::viridis(4, end = 0.8)[c(3, 1)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_fill",
values = viridis::viridis(4, end = 0.8)[c(3, 1)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_shape",
values = c(4, 3),
guide = "none") +
papaja::theme_apa() +
theme(legend.position = "bottom")
sitLie_trackgirl_diamonds_girl_hours <- sitLie_trackgirl_diamonds_girl %>%
dplyr::mutate_if(is.numeric, ~./60)
sitLie_trackgirl_diamonds_boy_hours <- sitLie_trackgirl_diamonds_boy %>%
dplyr::mutate_if(is.numeric, ~./60)
plot1 <- plot1 +
userfriendlyscience::diamondPlot(sitLie_trackgirl_diamonds_girl_hours,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[3],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15)) + # specify y-position; option to move the diamond
userfriendlyscience::diamondPlot(sitLie_trackgirl_diamonds_boy_hours,
returnLayerOnly = TRUE,
lineColor = "black",
linetype = "solid",
color=viridis::viridis(4, end = 0.8)[1],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15))
# Draw plot with intervention and control densities
plot2 <- df %>% dplyr::select(id,
track = track,
intervention,
PA = sitLieAccelerometer_T1) %>%
dplyr::filter(!is.na(track), track != "Other") %>% # Drop category "Other"
dplyr::mutate(track = factor(track, levels = c("IT", "BA", "HRC", "Nur")),
track = recode_factor(track, "IT" = "IT", "BA" = "BA",
"HRC" = "HRC", "Nur" = "Nur")) %>%
ggplot2::ggplot(aes(y = track)) +
ggridges::geom_density_ridges2(aes(x = PA, colour = "black",
fill = paste(track, intervention),
point_color = intervention,
point_fill = intervention,
point_shape = intervention),
scale = .75, alpha = 0.6, size = 0.25,
position = position_raincloud(width = 0.05, height = 0.15),
# from = 0, to = 450,
jittered_points = TRUE,
point_size = 1) +
labs(x = NULL,
y = NULL) +
scale_y_discrete(expand = c(0.1, 0), labels = NULL) +
scale_x_continuous(expand = c(0.01, 0)) +
ggridges::scale_fill_cyclical(
labels = c('BA 0' = "Control", 'BA 1' = "Intervention"),
values = viridis::viridis(4, end = 0.8)[c(2, 4)],
name = "",
guide = guide_legend(override.aes = list(alpha = 1,
point_shape = c(4, 3),
point_size = 2))) +
ggridges::scale_colour_cyclical(values = "black") +
ggridges::theme_ridges(grid = FALSE) +
# ggplot2::scale_fill_manual(values = c("#A0FFA0", "#A0A0FF")) +
ggridges::scale_discrete_manual(aesthetics = "point_color",
values = viridis::viridis(4, end = 0.8)[c(2, 4)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_fill",
values = viridis::viridis(4, end = 0.8)[c(2, 4)],
guide = "none") +
ggridges::scale_discrete_manual(aesthetics = "point_shape",
values = c(4, 3),
guide = "none") +
papaja::theme_apa() +
theme(legend.position = "bottom")
sitLie_trackintervention_diamonds_intervention_hours <- sitLie_trackintervention_diamonds_intervention %>%
dplyr::mutate_if(is.numeric, ~./60)
sitLie_trackintervention_diamonds_control_hours <- sitLie_trackintervention_diamonds_control %>%
dplyr::mutate_if(is.numeric, ~./60)
plot2 <- plot2 +
userfriendlyscience::diamondPlot(sitLie_trackintervention_diamonds_intervention,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[4],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15)) +
userfriendlyscience::diamondPlot(sitLie_trackintervention_diamonds_control,
returnLayerOnly = TRUE,
lineColor = "black",
color=viridis::viridis(4, end = 0.8)[2],
alpha=.6,
fixedSize = 0.1,
otherAxisCol = (1:4 + .15))
plot1 + plot2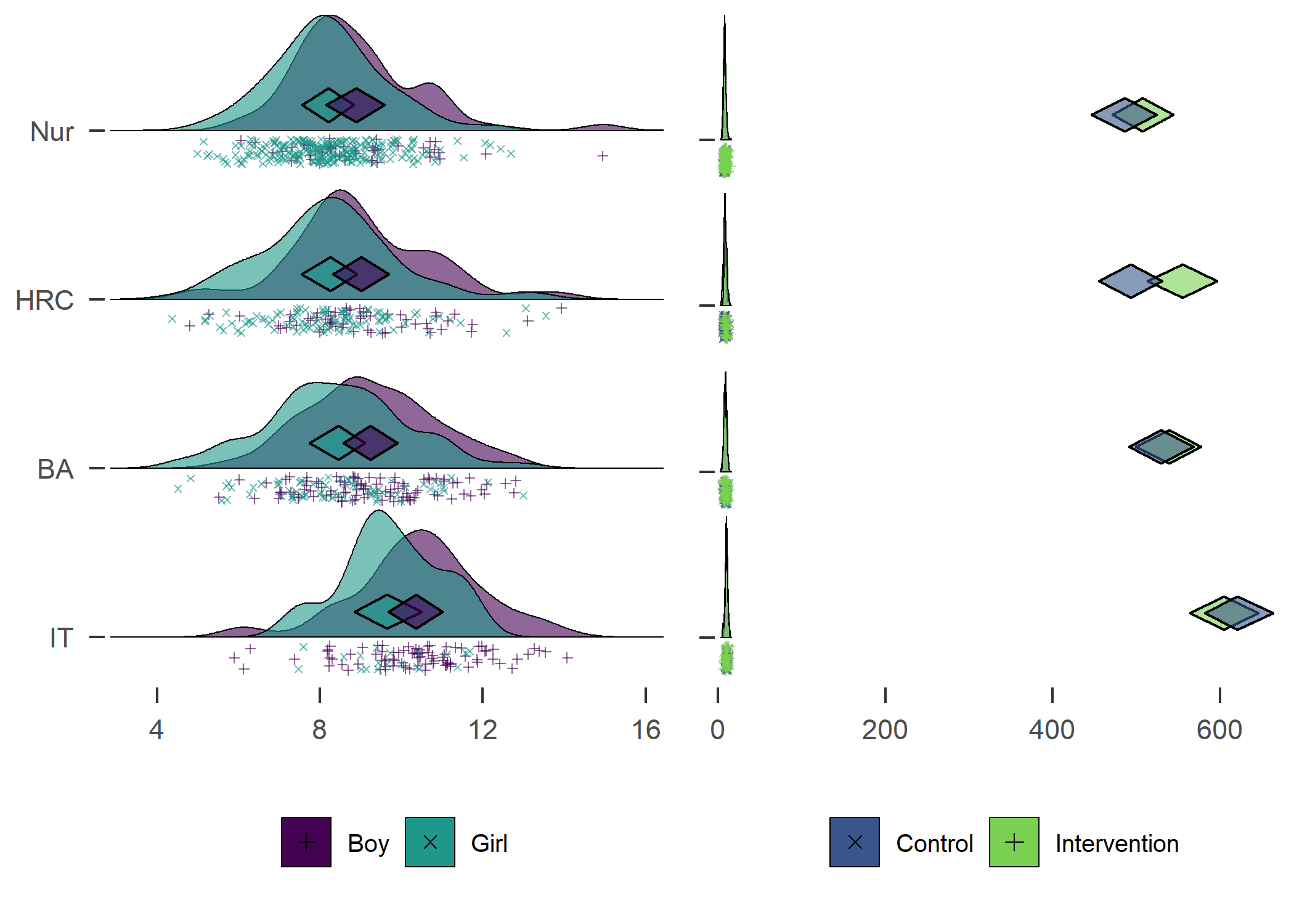
Average time spent sitting or lying down during a day. Nur = Practical nurse, HRC = Hotel, restaurant and catering, BA = Business and administration, IT = Information and communications technology.
Breaking the distributions down by tracks, this is what we see: boys are more sedentary in all groups, but the IT group–which consists of mostly boys–is also most sedentary regardless of gender. Distributions between intervention and control groups exhibit some differences, but the most pronounced one can be seen in the HRC track.